4MAP
 
 | | Crystal structure of Ara h 8 purified with heating | | Descriptor: | Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4M9B
 
 | | Crystal structure of Apo Ara h 8 | | Descriptor: | Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4M9W
 
 | | Crystal Structure of Ara h 8 with MES bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4MA6
 
 | | Crystal structure of Ara h 8 with Epicatechin bound | | Descriptor: | (2R,3R)-2-(3,4-dihydroxyphenyl)-3,4-dihydro-2H-chromene-3,5,7-triol, Ara h 8 allergen, SODIUM ION | | Authors: | Offermann, L.R, Hurlburt, B.K, Majorek, K.A, McBride, J.K, Maleki, S.J, Chruszcz, M. | | Deposit date: | 2013-08-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the Peanut Panallergen Ara h 8.
J.Biol.Chem., 288, 2013
|
|
4WOJ
 
 | | Aspartate Semialdehyde Dehydrogenase from Francisella tularensis | | Descriptor: | Aspartate semialdehyde dehydrogenase, SODIUM ION, SULFATE ION | | Authors: | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of aspartate b-semialdehyde dehydrogenase from Francisella tularensis
Acta Crystallogr.,Sect.F, 74, 2018
|
|
4TNN
 
 | | Crystal structure of Escherichia coli protein YodA in complex with Ni - artifact of purification. | | Descriptor: | Metal-binding lipocalin, NICKEL (II) ION, SULFATE ION | | Authors: | Gasiorowska, O.A, Cymborowski, M.T, Handing, K.B, Shabalin, I.G, Zasadzinska, E, Niedzialkowska, E, Porebski, P.J, Minor, W. | | Deposit date: | 2014-06-04 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
7KPS
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | Authors: | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-12 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
4ZNZ
 
 | | Crystal structure of Escherichia coli carbonic anhydrase (YadF) in complex with Zn - artifact of purification | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Gasiorowska, O.A, Niedzialkowska, E, Porebski, P.J, Handing, K.B, Shabalin, I.G, Cymborowski, M.T, Minor, W. | | Deposit date: | 2015-05-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
5HUO
 
 | | Crystal Structure of NadC Deletion Mutant in C2221 Space Group | | Descriptor: | Nicotinate-nucleotide diphosphorylase (Carboxylating), SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUL
 
 | | Crystal Structure of NadC Deletion Mutant in Cubic Space Group | | Descriptor: | PHOSPHATE ION, Quinolinate phosphoribosyltransferase | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.855 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUJ
 
 | | Crystal Structure of NadE from Streptococcus pyogenes | | Descriptor: | CHLORIDE ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5HUH
 
 | | Crystal Structure of NadE from Streptococcus pyogenes | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
3V48
 
 | | Crystal Structure of the putative alpha/beta hydrolase RutD from E.coli | | Descriptor: | GLYCEROL, Putative aminoacrylate hydrolase RutD, THIOCYANATE ION | | Authors: | Knapik, A.A, Petkowski, J.J, Otwinowski, Z, Cymborowski, M.T, Cooper, D.R, Chruszcz, M, Porebski, P.J, Niedzialkowska, E, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-04 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-faceted analysis of RutD reveals a novel family of alpha / beta hydrolases.
Proteins, 80, 2012
|
|
5HUP
 
 | | Crystal Structure of NadC from Streptococcus pyogenes | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase (Carboxylating), SULFATE ION | | Authors: | Booth, W.T, Chruszcz, M. | | Deposit date: | 2016-01-27 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Streptococcus pyogenes quinolinate-salvage pathway-structural and functional studies of quinolinate phosphoribosyl transferase and NH3 -dependent NAD(+) synthetase.
FEBS J., 284, 2017
|
|
5EM1
 
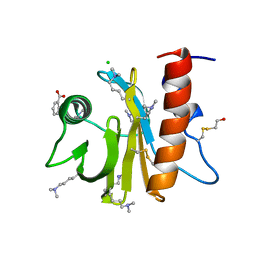 | | Crystal structure of ragweed allergen Amb a 8 | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Profilin | | Authors: | Offermann, L.R, He, J.Z, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
3V0R
 
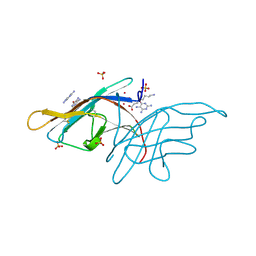 | | Crystal structure of Alternaria alternata allergen Alt a 1 | | Descriptor: | 2,5,6-triaminopyrimidin-4-ol, 8-aminooctanoic acid, Major allergen Alt a 1, ... | | Authors: | Chruszcz, M, Solberg, R, Osinski, T, Chapman, M.D, Minor, W. | | Deposit date: | 2011-12-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alternaria alternata allergen Alt a 1: a unique beta-barrel protein dimer found exclusively in fungi.
J.Allergy Clin.Immunol., 130, 2012
|
|
5EM0
 
 | | Crystal structure of mugwort allergen Art v 4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Pollen allergen Art v 4.01, SODIUM ION | | Authors: | Offermann, L.R, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-05 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
5EV0
 
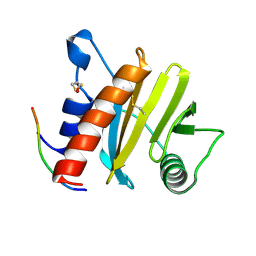 | | Crystal structure of ragweed profilin Amb a 8 in complex with poly-Pro14 | | Descriptor: | PRO-PRO-PRO-PRO-PRO-PRO-PRO-PRO-PRO, Profilin | | Authors: | Offermann, L.R, He, J.Z, Perdue, M.L, Chruszcz, M. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
5EVE
 
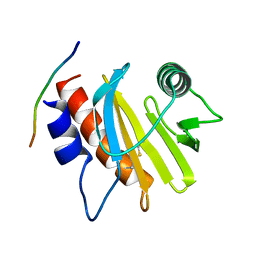 | | Crystal structure of Amb a 8 in complex with poly-Pro10 | | Descriptor: | Poly-Proline peptide, Profilin | | Authors: | Offermann, L.R, Schlachter, C.R, Garrett, J, Chruszcz, M. | | Deposit date: | 2015-11-19 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural, Functional, and Immunological Characterization of Profilin Panallergens Amb a 8, Art v 4, and Bet v 2.
J.Biol.Chem., 291, 2016
|
|
3RVT
 
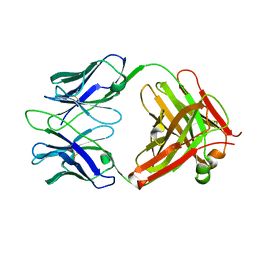 | | Structure of 4C1 Fab in P212121 space group | | Descriptor: | Fab fragment of 4C1 antibody - heavy chain, Fab fragment of 4C1 antibody - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
3RVU
 
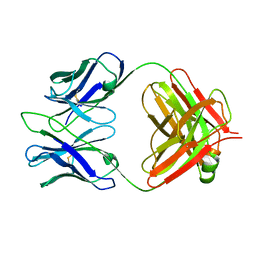 | | Structure of 4C1 Fab in C2221 space group | | Descriptor: | 4C1 Fab - heavy chain, 4C1 Fab - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
3S7I
 
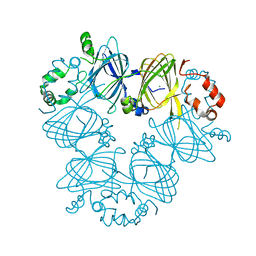 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
3S7E
 
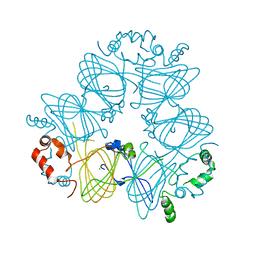 | | Crystal structure of Ara h 1 | | Descriptor: | Allergen Ara h 1, clone P41B, CHLORIDE ION | | Authors: | Chruszcz, M, Maleki, S.J, Solberg, R, Minor, W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Immunologic Characterization of Ara h 1, a Major Peanut Allergen.
J.Biol.Chem., 286, 2011
|
|
6BDX
 
 | | 4-hydroxy tetrahydrodipicolinate reductase from Neisseria gonorrhoeae | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, SULFATE ION | | Authors: | Pote, S.S, Pye, S.E, Sheahan, T.E, Chruszcz, M. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Hydroxy-tetrahydrodipicolinate reductase from Neisseria gonorrhoeae - structure and interactions with coenzymes and substrate analog.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
