5JLS
 
 | |
5JLU
 
 | |
6O5C
 
 | | X-ray crystal structure of metal-dependent transcriptional regulator MtsR | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Putative metal-dependent transcriptional regulator | | Authors: | Do, H, Kumaraswami, M. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Metal sensing and regulation of adaptive responses to manganese limitation by MtsR is critical for group A streptococcus virulence.
Nucleic Acids Res., 47, 2019
|
|
6OF0
 
 | | Structural basis for multidrug recognition and antimicrobial resistance by MTRR, an efflux pump regulator from Neisseria Gonorrhoeae | | Descriptor: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Beggs, G.A, Kumaraswami, M, Shafer, W, Brennan, R.G. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and In Vivo Characterization of MtrR-Mediated Resistance to Innate Antimicrobials by the Human Pathogen Neisseria gonorrhoeae .
J.Bacteriol., 201, 2019
|
|
3HSY
 
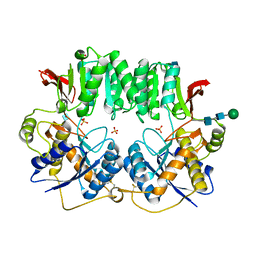 | | High resolution structure of a dimeric GluR2 N-terminal domain (NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2, SULFATE ION, ... | | Authors: | Rossmann, M, Sukumaran, M, Penn, A.C, Veprintsev, D.B, Greger, I.H. | | Deposit date: | 2009-06-11 | | Release date: | 2010-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Subunit-selective N-terminal domain associations organize the formation of AMPA receptor heteromers
Embo J., 30, 2011
|
|
6DQL
 
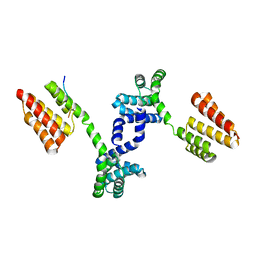 | | Crystal structure of Regulator of Proteinase B RopB complexed with SIP | | Descriptor: | Regulator of Proteinase B RopB, SpeB-inducing peptide (SIP) | | Authors: | Do, H, Makthal, N, VanderWal, A.R, Olsen, R.J, Musser, J.M, Kumaraswami, M. | | Deposit date: | 2018-06-11 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway.
Nat Commun, 10, 2019
|
|
2Y8P
 
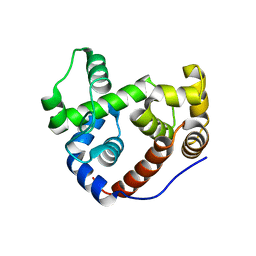 | | Crystal Structure of an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase (MltE) from Escherichia coli | | Descriptor: | ENDO-TYPE MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE A | | Authors: | Artola-Recolons, C, Carrasco-Lopez, C, Llarrull, L.I, Kumarasiri, M, Lastochkin, E, Martinez-Ilarduya, I, Meindl, K, Uson, I, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2011-02-08 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | High-Resolution Crystal Structure of Mlte, an Outer Membrane-Anchored Endolytic Peptidoglycan Lytic Transglycosylase from Escherichia Coli.
Biochemistry, 50, 2011
|
|
6K5Q
 
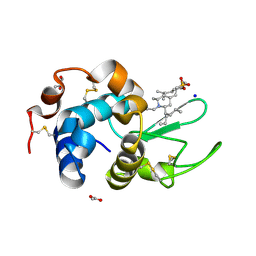 | | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia | | Descriptor: | ACETATE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Kanal Elamparithi, B, Ankur, T, Sivakumar, M, Gunasekaran, K. | | Deposit date: | 2019-05-30 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.177 Å) | | Cite: | Crystal structure of lysozyme complexed with a bioactive compound from Jatropha gossypiifolia
To Be Published
|
|
7NVJ
 
 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NW1
 
 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
6PU8
 
 | | Room temperature X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate of keto-darunavir | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3,3-dihydroxy-1-phenylbutan-2-yl]carbamate, HIV-1 protease | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-17 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
6PTP
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | HIV-1 Protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Kovalevsky, A, Das, A. | | Deposit date: | 2019-07-16 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
6KMP
 
 | | 100K X-ray structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with tetrahedral intermediate mimic KVS-1 | | Descriptor: | N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide, Protease | | Authors: | Das, A, Kovalevsky, A. | | Deposit date: | 2019-07-31 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Visualizing Tetrahedral Oxyanion Bound in HIV-1 Protease Using Neutrons: Implications for the Catalytic Mechanism and Drug Design.
Acs Omega, 5, 2020
|
|
3UY6
 
 | |
5ZHV
 
 | | Crystal structure of the PadR-family transcriptional regulator Rv3488 of Mycobacterium tuberculosis H37Rv in complex with zinc ion | | Descriptor: | Transcriptional regulator, ZINC ION | | Authors: | Meera, K, Pal, R.K, Arora, A, Biswal, B.K. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of the transcriptional regulator Rv3488 ofMycobacterium tuberculosisH37Rv.
Biochem. J., 475, 2018
|
|
5ZHC
 
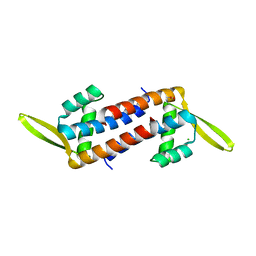 | | Crystal structure of the PadR-family transcriptional regulator Rv3488 of Mycobacterium tuberculosis H37Rv | | Descriptor: | ACETATE ION, CHLORIDE ION, Transcriptional regulator | | Authors: | Meera, K, Pal, R.K, Arora, A, Biswal, B.K. | | Deposit date: | 2018-03-12 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterization of the transcriptional regulator Rv3488 ofMycobacterium tuberculosisH37Rv.
Biochem. J., 475, 2018
|
|
5ZI8
 
 | | Crystal structure of the PadR-family transcriptional regulator Rv3488 of Mycobacterium tuberculosis H37Rv in complex with cadmium ion | | Descriptor: | CADMIUM ION, Transcriptional regulator | | Authors: | Meera, K, pal, R.K, Arora, A, Biswal, B.K. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of the transcriptional regulator Rv3488 ofMycobacterium tuberculosisH37Rv.
Biochem. J., 475, 2018
|
|
5J61
 
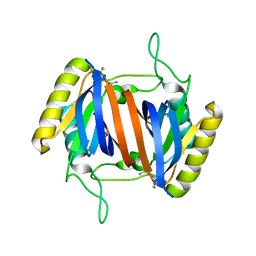 | |
3PUO
 
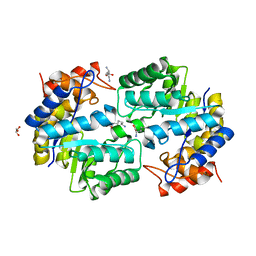 | | Crystal structure of dihydrodipicolinate synthase from Pseudomonas aeruginosa(PsDHDPS)complexed with L-lysine at 2.65A resolution | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, LYSINE | | Authors: | Kaur, N, Kumar, M, Kumar, S, Gautam, A, Sinha, M, Kaur, P, Sharma, S, Sharma, R, Tewari, R, Singh, T.P. | | Deposit date: | 2010-12-06 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical studies and crystal structure determination of dihydrodipicolinate synthase from Pseudomonas aeruginosa
Int.J.Biol.Macromol., 48, 2011
|
|
4U4M
 
 | | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate | | Descriptor: | 1,2-ETHANEDIOL, PYRUVIC ACID, UREA, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of 0.5M urea unfolded YagE, a KDG aldolase protein in complex with Pyruvate
To be published
|
|
3N2X
 
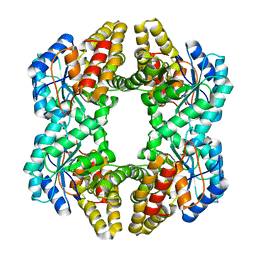 | | Crystal structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 in complex with pyruvate | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-05-19 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
3NEV
 
 | | Crystal structure of YagE, a prophage protein from E. coli K12 in complex with KDGal | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-LYXO-HEXONIC ACID, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
8FCB
 
 | | Cryo-EM structure of the human TRPV4 - RhoA in complex with GSK1016790A | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHOLESTEROL HEMISUCCINATE, N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, ... | | Authors: | Kwon, D.H, Lee, S.-Y, Zhang, F. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | TRPV4-Rho GTPase complex structures reveal mechanisms of gating and disease.
Nat Commun, 14, 2023
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
