5BXP
 
 | | LNBase in complex with LNB-LOGNAc | | Descriptor: | Lacto-N-biosidase, SULFATE ION, beta-D-galactopyranose-(1-3)-N-acetylglucosaminono-1,5-lactone (Z)-oxime | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXS
 
 | | LNBase in complex with LNB-NHAcCAS | | Descriptor: | 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXR
 
 | | LNBase in complex with LNB-NHAcDNJ | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Lacto-N-biosidase, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
5BXT
 
 | | LNBase in complex with LNB-NHAcAUS | | Descriptor: | Lacto-N-biosidase, N-{[(1R,2R,3R,7S,7aR)-1,2,7-trihydroxyhexahydro-1H-pyrrolizin-3-yl]methyl}acetamide, SULFATE ION, ... | | Authors: | Ito, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the catalysis by GH20 lacto-N-biosidase using small molecule inhibitors and structural analysis
Chem.Commun.(Camb.), 51, 2015
|
|
6KQT
 
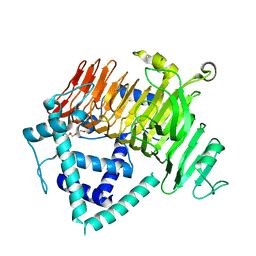 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
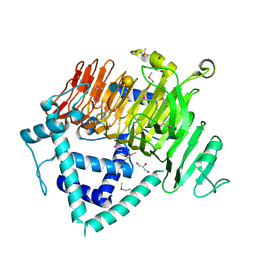 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
5WZN
 
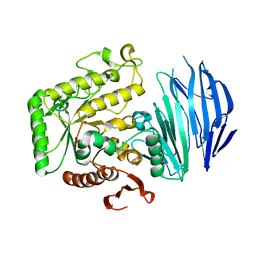 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
6HUS
 
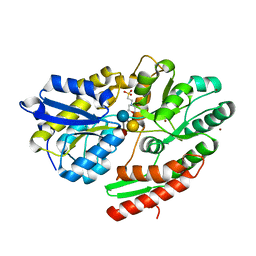 | | 2'-fucosyllactose and 3-fucosyllactose binding protein from Bifidobacterium longum infantis, bound with 3-fucosyllactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ABC transporter substrate-binding protein, ZINC ION, ... | | Authors: | Ejby, M, Abou Hachem, M, Lo Leggio, L, Takane, K, Sakanaka, M. | | Deposit date: | 2018-10-09 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis.
Sci Adv, 5, 2019
|
|
5WQV
 
 | | Crystal structure of PriB mutant - S55A | | Descriptor: | Primosomal replication protein N | | Authors: | Fujiyama, S, Shiroishi, M, Katayama, T, Abe, Y, Ueda, T. | | Deposit date: | 2016-11-28 | | Release date: | 2017-11-29 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Insight into the interaction between PriB and DnaT on bacterial DNA replication restart: Significance of the residues on PriB dimer interface and highly acidic region on DnaT.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
7V6M
 
 | |
7V6I
 
 | |
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
