6X3C
 
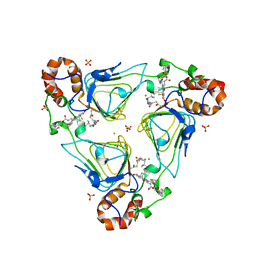 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F1037 (47) | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
6X3J
 
 | | Crystal structure of streptogramin A acetyltransferase VatA from Staphylococcus aureus in complex with streptogramin analog F0224 (46) | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Chaires, H.A, Fraser, J.S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
2PMQ
 
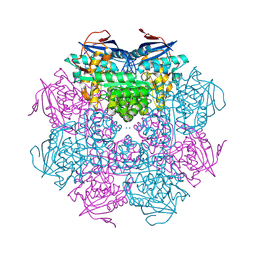 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme from Roseovarius sp. HTCC2601 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Sridhar, V, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-04-23 | | Release date: | 2007-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of new enzymes and metabolic pathways by using structure and genome context.
Nature, 502, 2013
|
|
3ORZ
 
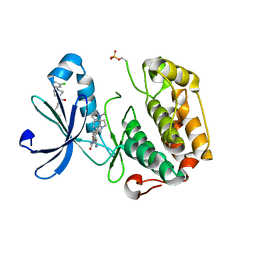 | | PDK1 mutant bound to allosteric disulfide fragment activator 2A2 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(3-chlorophenyl)piperazin-1-yl]-4-oxobutane-1-thiol | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9995 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ORX
 
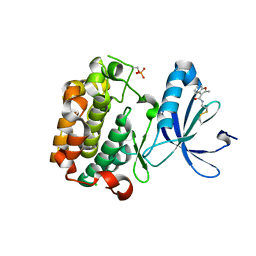 | | PDK1 mutant bound to allosteric disulfide fragment inhibitor 1F8 | | Descriptor: | 2-methyl-N-(2-sulfanylethyl)-1-benzofuran-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, CHLORIDE ION | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2044 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P41
 
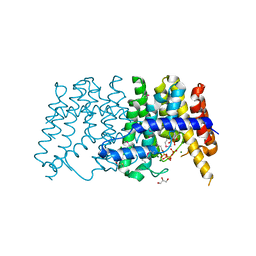 | | Crystal structure of polyprenyl synthetase from pseudomonas fluorescens pf-5 complexed with magnesium and isoprenyl pyrophosphate | | Descriptor: | CHLORIDE ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-05 | | Release date: | 2010-10-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2RB1
 
 | |
2RBT
 
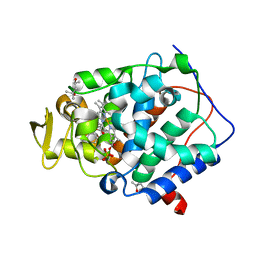 | | n-methylbenzylamine in complex with Cytochrome C Peroxidase W191G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cytochrome C Peroxidase, N-methyl-1-phenylmethanamine, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBX
 
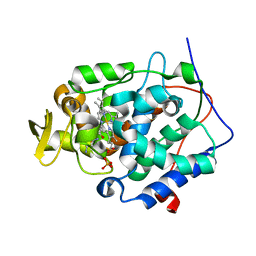 | | Cytochrome C Peroxidase W191G in complex with pyrimidine-2,4,6-triamine. | | Descriptor: | Cytochrome C Peroxidase, PHOSPHATE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBW
 
 | | Cytochrome C Peroxidase W191G in complex with 1,2-dimethyl-1h-pyridin-5-amine | | Descriptor: | 5-amino-1,2-dimethylpyridinium, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
2RBN
 
 | |
2RBZ
 
 | | Cytochrome C Peroxidase W191G in complex 3-methoxypyridine | | Descriptor: | 3-methoxypyridine, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
3PDE
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Lactobacillus brevis atcc 367 complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Farnesyl-diphosphate synthase, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-22 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q4D
 
 | | Crystal structure of dipeptide epimerase from Cytophaga hutchinsonii complexed with Mg and dipeptide D-Ala-L-Ala | | Descriptor: | ALANINE, D-ALANINE, MAGNESIUM ION, ... | | Authors: | Lukk, T, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2010-12-23 | | Release date: | 2011-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q2Q
 
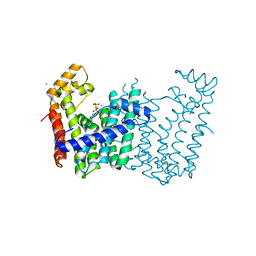 | | Crystal structure of geranylgeranyl pyrophosphate synthase from Corynebacterium glutamicum complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3QQV
 
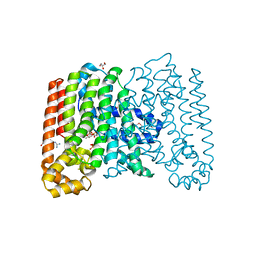 | | Crystal structure of geranylgeranyl pyrophosphate synthase from corynebacterium glutamicum complexed with isoprenyl diphosphate and magnesium | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, GLYCEROL, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-02-16 | | Release date: | 2011-03-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3R0U
 
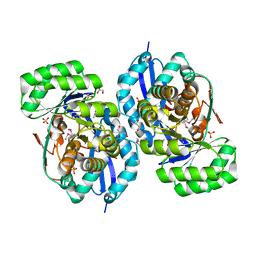 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Tartrate and Mg complex | | Descriptor: | D(-)-TARTARIC ACID, Enzyme of enolase superfamily, GLYCEROL, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3PKO
 
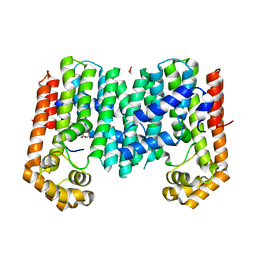 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 complexed with citrate | | Descriptor: | CITRIC ACID, GLYCEROL, Geranylgeranyl pyrophosphate synthase | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Poulter, C.D, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-11-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3Q1O
 
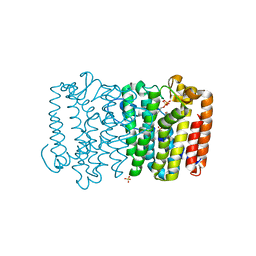 | | Crystal structure of geranyltransferase from helicobacter pylori complexed with magnesium and isoprenyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Geranyltranstransferase (IspA), MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3P8R
 
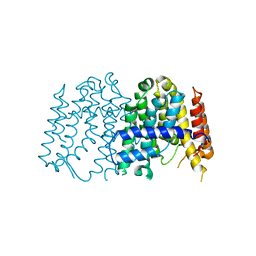 | | CRYSTAL STRUCTURE OF POLYPRENYL SYNTHASE FROM Vibrio cholerae | | Descriptor: | Geranyltranstransferase | | Authors: | Patskovsky, Y, Toro, R, Dickey, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3RIT
 
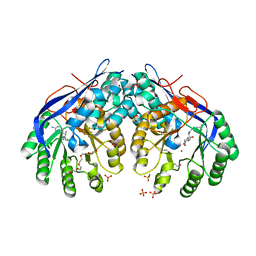 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg and dipeptide L-Arg-D-Lys | | Descriptor: | ARGININE, D-LYSINE, Dipeptide epimerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OTU
 
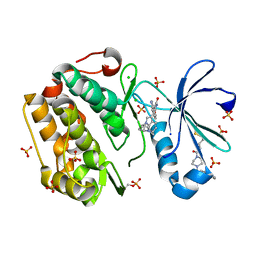 | | PDK1 mutant bound to allosteric disulfide fragment activator JS30 | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, 3-phosphoinositide-dependent protein kinase 1, 4-[4-(naphthalen-1-ylmethyl)piperazin-1-yl]-4-oxobutane-1-thiol, ... | | Authors: | Sadowsky, J.D, Wells, J.A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1013 Å) | | Cite: | Turning a protein kinase on or off from a single allosteric site via disulfide trapping.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OYR
 
 | | Crystal structure of polyprenyl synthase from Caulobacter crescentus CB15 complexed with calcium and isoprenyl diphosphate | | Descriptor: | CALCIUM ION, DIMETHYLALLYL DIPHOSPHATE, PYROPHOSPHATE 2-, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Poulter, C.D, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-09-23 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3R0K
 
 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Tartrate bound, no Mg | | Descriptor: | D(-)-TARTARIC ACID, Enzyme of enolase superfamily, GLYCEROL, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RMG
 
 | | Crystal structure of geranylgeranyl pyrophosphate synthase from bacteroides thetaiotaomicron | | Descriptor: | Octaprenyl-diphosphate synthase | | Authors: | Patskovsky, Y, Toro, R, Sauder, J.M, Poulter, C.D, Gerlt, J.A, Burley, S.K, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-04-20 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prediction of function for the polyprenyl transferase subgroup in the isoprenoid synthase superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
