1WSW
 
 | | Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, dimer, semiquinone state | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Redox properties and crystal structures of a Desulfovibrio vulgaris flavodoxin mutant in the monomeric and homodimeric forms.
Biochim.Biophys.Acta, 1794, 2009
|
|
3WNO
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltooctaose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
1XT6
 
 | | S35C Flavodoxin Mutant in the semiquinone state | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Cassetta, A, Sassone, C, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-10-21 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of S35C flavodoxin mutant from Desulfovibrio vulgaris in the semiquinone state.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1XYV
 
 | | Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, monomer, semiquinone state | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-11-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Redox properties and crystal structures of a Desulfovibrio vulgaris flavodoxin mutant in the monomeric and homodimeric forms.
Biochim.Biophys.Acta, 1794, 2009
|
|
3WNL
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNM
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNK
 
 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | Descriptor: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
1V6Q
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.3 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1XYY
 
 | | Low Temperature (100K) Crystal Structure Of Flavodoxin Mutant S64C, homodimer, oxidised state | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Artali, R, Marchini, N, Meneghetti, F, Cavazzini, D, Bombieri, G, Rossi, G.L, Gilardi, G. | | Deposit date: | 2004-11-11 | | Release date: | 2004-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Redox properties and crystal structures of a Desulfovibrio vulgaris flavodoxin mutant in the monomeric and homodimeric forms.
Biochim.Biophys.Acta, 1794, 2009
|
|
1V4F
 
 | | Crystal structures of collagen model peptides with pro-hyp-gly sequence at 1.3A | | Descriptor: | collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-11-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V7H
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
3WNN
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3AH9
 
 | | Crystal structure of (Pro-Pro-Gly)9 at 1.1 A resolution | | Descriptor: | ACETIC ACID, AZIDE ION, collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Hosaka, N, Nishino, N. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of (Pro-Pro-Gly)9
To be Published
|
|
1FFG
 
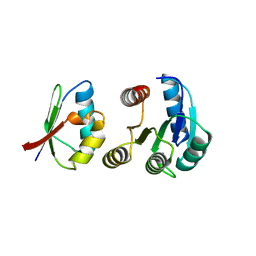 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY AT 2.1 A RESOLUTION | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFW
 
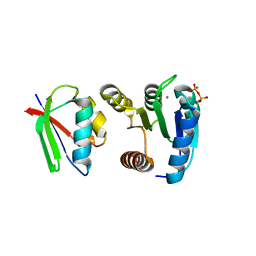 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY WITH A BOUND IMIDO DIPHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, IMIDO DIPHOSPHATE, ... | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1FFS
 
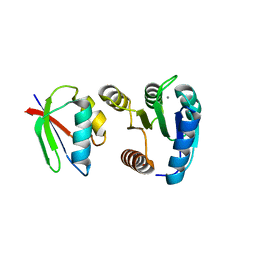 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7AJR
 
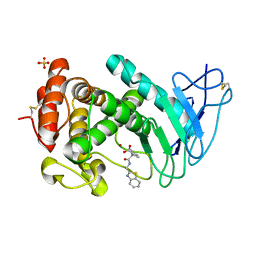 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
4LNK
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | Descriptor: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5793 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNO
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: form two of GS-1 | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNF
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-Q | | Descriptor: | GLUTAMINE, Glutamine synthetase, MAGNESIUM ION, ... | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
7BYT
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYX
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208A with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYS
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
