6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
5BS2
 
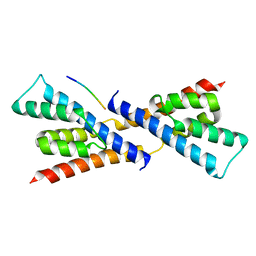 | | Crystal structure of RbcX-IIa from Chlamydomonas reinhardtii in complex with RbcL C-terminal tail | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase large chain,CrRbcX-IIa | | Authors: | Bracher, A, Hauser, T, Liu, C, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of the Rubisco-Assembly Chaperone RbcX-II from Chlamydomonas reinhardtii.
Plos One, 10, 2015
|
|
5D5V
 
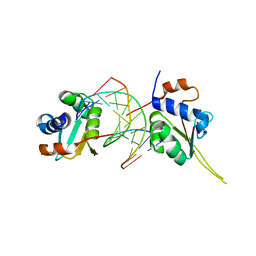 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5U
 
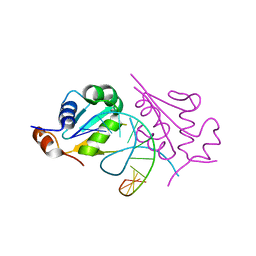 | | Crystal structure of human Hsf1 with HSE DNA | | Descriptor: | Heat shock Element DNA, Heat shock factor protein 1 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D60
 
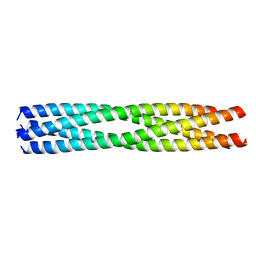 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form III | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5X
 
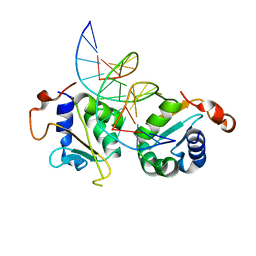 | | Crystal structure of Chaetomium thermophilum Skn7 with SSRE DNA | | Descriptor: | Putative transcription factor, SSRE DNA strand 1, SSRE DNA strand 2 | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5W
 
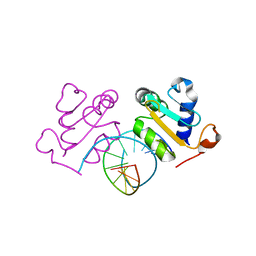 | | Crystal structure of Chaetomium thermophilum Skn7 with HSE DNA | | Descriptor: | HSE DNA, Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Z
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form II | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Bracher, A, Hartl, F.U. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5D5Y
 
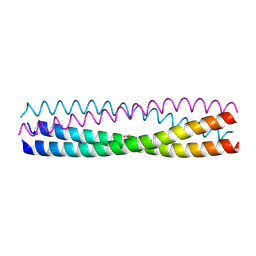 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6HB1
 
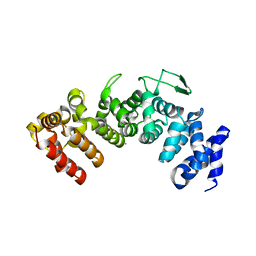 | | Structure of Hgh1, crystal form I | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB3
 
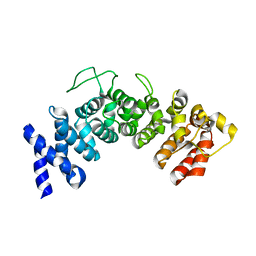 | | Structure of Hgh1, crystal form II | | Descriptor: | Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB2
 
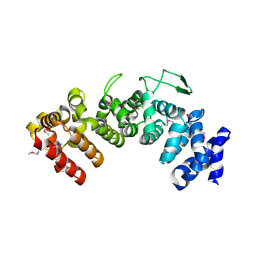 | | Structure of Hgh1, crystal form I, Selenomethionine derivative | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
1HDJ
 
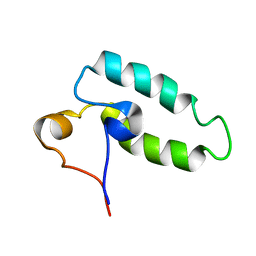 | | HUMAN HSP40 (HDJ-1), NMR | | Descriptor: | HUMAN HSP40 | | Authors: | Qian, Y.Q, Patel, D, Hartl, F.-U, Mccoll, D.J. | | Deposit date: | 1996-05-09 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain.
J.Mol.Biol., 260, 1996
|
|
3HYB
 
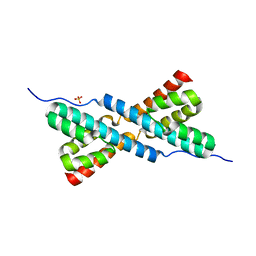 | |
1HX1
 
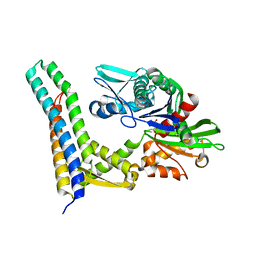 | | CRYSTAL STRUCTURE OF A BAG DOMAIN IN COMPLEX WITH THE HSC70 ATPASE DOMAIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG family molecular chaperone regulator 1, Heat shock 70 kDa protein 8 | | Authors: | Sondermann, H, Scheufler, C, Moarefi, I. | | Deposit date: | 2001-01-11 | | Release date: | 2001-03-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors.
Science, 291, 2001
|
|
1FXK
 
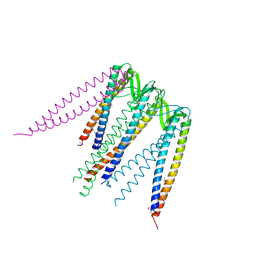 | |
3D2E
 
 | | Crystal structure of a complex of Sse1p and Hsp70, Selenomethionine-labeled crystals | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
3D2F
 
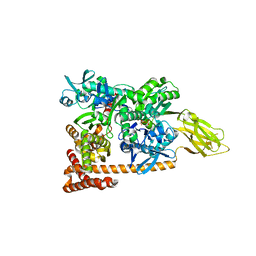 | | Crystal structure of a complex of Sse1p and Hsp70 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
6Z1E
 
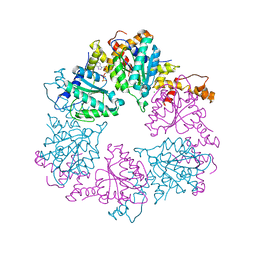 | |
6Z1D
 
 | |
6R86
 
 | | Yeast Vms1-60S ribosomal subunit complex (post-state) | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R87
 
 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R84
 
 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state with Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
