5MBC
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBB
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBH
 
 | | Structure of a bacterial light-regulated adenylyl cyclase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBK
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Beta subunit of photoactivated adenylyl cyclase, MAGNESIUM ION | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5M2A
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5M27
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-10-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBJ
 
 | | Structure of a bacterial light-regulated adenylyl cyclase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
5MBE
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
7ABR
 
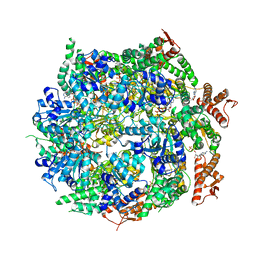 | | Cryo-EM structure of B. subtilis ClpC (DWB mutant) hexamer bound to a substrate polypeptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Negative regulator of genetic competence ClpC/MecB, ... | | Authors: | Morreale, F.E, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2020-09-08 | | Release date: | 2021-10-06 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | BacPROTACs mediate targeted protein degradation in bacteria.
Cell, 185, 2022
|
|
8BRG
 
 | |
7OYB
 
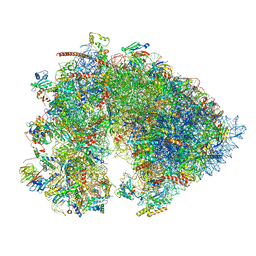 | | Cryo-EM structure of the 6 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYA
 
 | | Cryo-EM structure of the 1 hpf zebrafish embryo 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYD
 
 | | Cryo-EM structure of a rabbit 80S ribosome with zebrafish Dap1b | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
7OYC
 
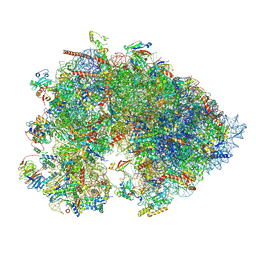 | | Cryo-EM structure of the Xenopus egg 80S ribosome | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Leesch, F, Lorenzo-Orts, L, Grishkovskaya, I, Kandolf, S, Belacic, K, Meinhart, A, Haselbach, D, Pauli, A. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-20 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A molecular network of conserved factors keeps ribosomes dormant in the egg.
Nature, 613, 2023
|
|
1J98
 
 | | The 1.2 Angstrom Structure of Bacillus subtilis LuxS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.2 A Structure of a Novel Quorum-Sensing Protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
5FGT
 
 | | Thaumatin solved by native sulphur-SAD using free-electron laser radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.K, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
5FGX
 
 | | Thaumatin solved by native sulphur SAD using synchrotron radiation | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K.J, Meinhart, A, Barends, T.R.M, Foucar, L, Gorel, A, Aquila, A, Botha, S, Doak, R.B, Koglin, J, Liang, M, Shoeman, R.L, Williams, G.J, Boutet, S, Schlichting, I. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data.
Iucrj, 3, 2016
|
|
1JQW
 
 | | THE 2.3 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/HOMOCYSTEINE COMPLEX | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ZINC ION | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-09 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
1JVI
 
 | | THE 2.2 ANGSTROM RESOLUTION STRUCTURE OF BACILLUS SUBTILIS LUXS/RIBOSILHOMOCYSTEINE COMPLEX | | Descriptor: | (2S)-2-amino-4-[[(2S,3S,4R,5R)-3,4,5-trihydroxyoxolan-2-yl]methylsulfanyl]butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, Autoinducer-2 production protein luxS, ... | | Authors: | Ruzheinikov, S.N, Das, S.K, Sedelnikova, S.E, Hartley, A, Foster, S.J, Horsburgh, M.J, Cox, A.G, McCleod, C.W, Mekhalfia, A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-08-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.2 A structure of a novel quorum-sensing protein, Bacillus subtilis LuxS
J.Mol.Biol., 313, 2001
|
|
7QVE
 
 | | Spinach 20S proteasome | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit alpha type-3, Proteasome subunit beta, ... | | Authors: | Kandolf, S, Grishkovskaya, I, Meinhart, A, Haselbach, D. | | Deposit date: | 2022-01-21 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the plant 26S proteasome.
Plant Commun., 3, 2022
|
|
5NBY
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2017-03-02 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
4OI1
 
 | | Clp1 bound to ssRNA dinucleotide GC, ADP, AlF4-, and Mg2+(transition state, data set II) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
4OI2
 
 | | C. Elegans Clp1 and ADP and Mg2+ (turnover state) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NONAETHYLENE GLYCOL, ... | | Authors: | Dikfidan, A, Loll, B, Zeymer, C, Clausen, T, Meinhart, A. | | Deposit date: | 2014-01-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase clp1.
Mol.Cell, 54, 2014
|
|
2BYM
 
 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CADMIUM ION, CHRAC-14, CHRAC-16 | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
2BYK
 
 | | Histone fold heterodimer of the Chromatin Accessibility Complex | | Descriptor: | CHRAC-14, CHRAC-16, SULFATE ION | | Authors: | Fernandez-Tornero, C, Hartlepp, K.F, Grune, T, Eberharter, A, Becker, P.B, Muller, C.W. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Histone Fold Subunits of Drosophila Chrac Facilitate Nucleosome Sliding Through Dynamic DNA Interactions.
Mol.Cell.Biol., 25, 2005
|
|
