4A22
 
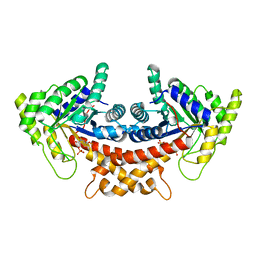 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to N-(4-hydroxybutyl)- glycolohydroxamic acid bis- phosphate | | Descriptor: | 4-{hydroxy[(phosphonooxy)acetyl]amino}butyl dihydrogen phosphate, FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, ... | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycolytic and non-glycolytic functions of Mycobacterium tuberculosis fructose-1,6-bisphosphate aldolase, an essential enzyme produced by replicating and non-replicating bacilli.
J. Biol. Chem., 286, 2011
|
|
4A0Y
 
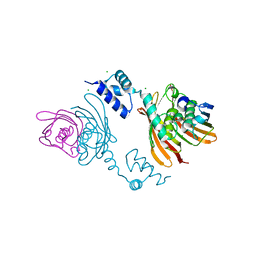 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4A0Z
 
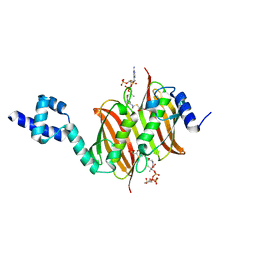 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with malonyl-CoA | | Descriptor: | MALONYL-COENZYME A, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4A12
 
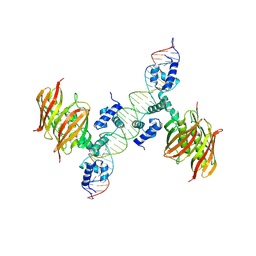 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with DNA operator | | Descriptor: | FAPR PROMOTER, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4A21
 
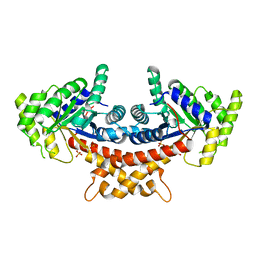 | | Structure of Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase bound to sulfate | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE, SODIUM ION, SULFATE ION | | Authors: | Coincon, M, De la Paz Santangelo, M, Gest, P.M, Guerin, M.E, Pham, H, Ryan, G, Puckett, S.E, Spencer, J.S, Gonzalez-Juarrero, M, Daher, R, Lenaerts, A.J, Schnappinger, D, Therisod, M, Ehrt, S, Jackson, M, Sygusch, J. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glycolytic and Non-Glycolytic Functions of the Fructose-1,6-Bisphosphate Aldolase of Mycobacterium Tuberculosis, an Essential Enzyme Produced by Replicating and Non-Replicating Bacilli
J.Biol.Chem., 286, 2011
|
|
4DE7
 
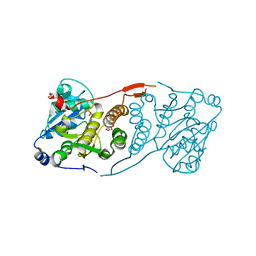 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mg2+ and uridine-diphosphate (UDP) | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4DDZ
 
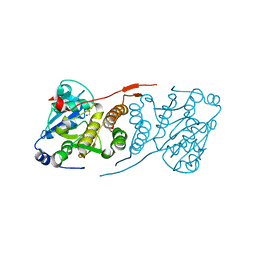 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis | | Descriptor: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL | | Authors: | Albesa-Jove, D, Urresti, S, Gest, P.M, van der Woerd, M, Jackson, M, Guerin, M.E. | | Deposit date: | 2012-01-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
6R8B
 
 | | Escherichia coli AGPase in complex with FBP. | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-01 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6R8U
 
 | | Escherichia coli AGPase in complex with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-04-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM
Biorxiv, 2020
|
|
6T8K
 
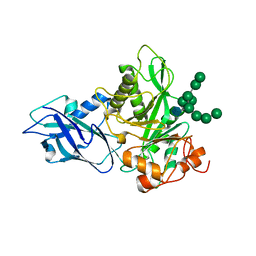 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc product in P1 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6TCV
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 in complex with Man9GlcNAc2Asn substrate | | Descriptor: | ASPARAGINE, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8I
 
 | | Crystal structure of wild type EndoBT-3987 from Bacteroides thetaiotamicron VPI-5482 | | Descriptor: | Endo-beta-N-acetylglucosaminidase F1, GLYCEROL | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6T8L
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man9GlcNAc product in P212121 | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-10-24 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6TCW
 
 | | Crystal structure of Bacteroides thetaiotamicron EndoBT-3987 with Man5GlcNAc product | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F1, GLYCEROL, ... | | Authors: | Trastoy, B, Du, J.J, Klontz, E.H, Cifuente, J.O, Sundberg, E.J, Guerin, M.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structural basis of mammalian high-mannose N-glycan processing by human gut Bacteroides.
Nat Commun, 11, 2020
|
|
6MDV
 
 | | Crystal structure of Streptococcus pyogenes endo-beta-N-acetylglucosaminidase (EndoS2) with high-mannose glycan | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klontz, E.H, Trastoy, B, Orwenyo, J, Wang, L.X, Guerin, M.E, Sundberg, E.J. | | Deposit date: | 2018-09-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Basis of Broad SpectrumN-Glycan Specificity and Processing of Therapeutic IgG Monoclonal Antibodies by Endoglycosidase S2.
ACS Cent Sci, 5, 2019
|
|
1RZU
 
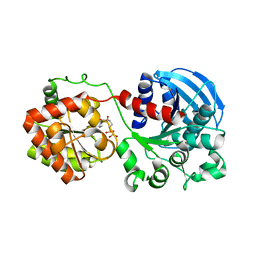 | | Crystal structure of the glycogen synthase from A. tumefaciens in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase 1 | | Authors: | Buschiazzo, A, Guerin, M.E, Ugalde, J.E, Ugalde, R.A, Shepard, W, Alzari, P.M. | | Deposit date: | 2003-12-29 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation.
Embo J., 23, 2004
|
|
1RZV
 
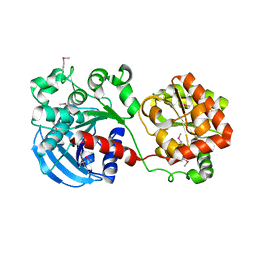 | | Crystal structure of the glycogen synthase from Agrobacterium tumefaciens (non-complexed form) | | Descriptor: | Glycogen synthase 1 | | Authors: | Buschiazzo, A, Guerin, M.E, Ugalde, J.E, Ugalde, R.A, Shepard, W, Alzari, P.M. | | Deposit date: | 2003-12-29 | | Release date: | 2004-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation.
Embo J., 23, 2004
|
|
4DEC
 
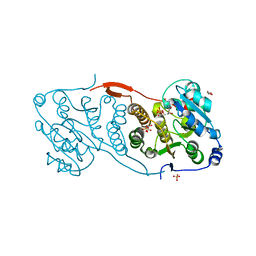 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate (UDP) and phosphoglyceric acid (PGA) | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL, ... | | Authors: | Albesa-Jove, D, Urresti, S, van der Woerd, M, Guerin, M.E. | | Deposit date: | 2012-01-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4NYY
 
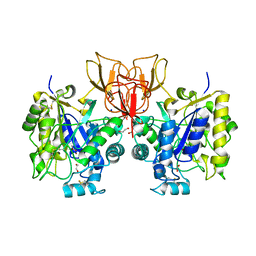 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with acetate ion (ACT) in P 2 21 21 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ1
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ4
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with 2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE (NDG) and zinc ion | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NXI
 
 | | Rv2466c Mediates the Activation of TP053 To Kill Replicating and Non-replicating Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Guerin, M.E. | | Deposit date: | 2013-12-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Rv2466c mediates the activation of TP053 to kill replicating and non-replicating Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
4NZ3
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NZ5
 
 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with 2-(ACETYLAMINO)-2-DEOXY-A-D-GLUCOPYRANOSE (NDG) and cadmium ion | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, ACETATE ION, CADMIUM ION, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Vibrio cholerae protein
To be Published
|
|
4N9W
 
 | | Crystal structure of phosphatidyl mannosyltransferase PimA | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose-dependent alpha-(1-2)-phosphatidylinositol mannosyltransferase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Giganti, D, Albesa-Jove, D, Bellinzoni, M, Guerin, M.E, Alzari, P.M. | | Deposit date: | 2013-10-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Secondary structure reshuffling modulates glycosyltransferase function at the membrane.
Nat.Chem.Biol., 11, 2015
|
|
