4AIA
 
 | |
3QSI
 
 | |
3PHT
 
 | |
4NUY
 
 | | Crystal structure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NUZ
 
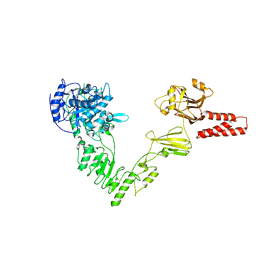 | | Crystal structure of a glycosynthase mutant (D233Q) of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2W02
 
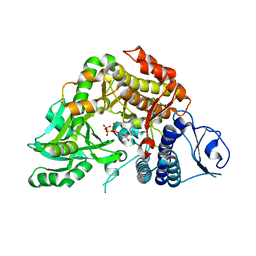 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with ATP from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
Nat. Chem. Biol., 5, 2009
|
|
2W0M
 
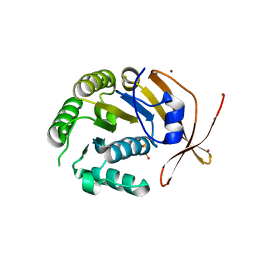 | | Crystal Structure of sso2452 from Sulfolobus solfataricus P2 | | Descriptor: | PYROPHOSPHATE 2-, SSO2452, ZINC ION | | Authors: | McRobbie, A, Carter, L, Johnson, K.A, Kerou, M, Liu, H, Mcmahon, S, Oke, M, Naismith, J.H, White, M.F. | | Deposit date: | 2008-08-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterisation of a Conserved Archaeal Rada Paralog with Antirecombinase Activity.
J.Mol.Biol., 389, 2009
|
|
2X5Q
 
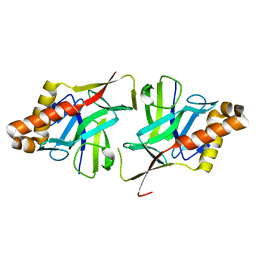 | | Crystal Structure of Hypothetical protein sso1986 from Sulfolobus solfataricus P2 | | Descriptor: | SSO1986 | | Authors: | Oke, M, Carter, L, Johnson, K.A, Kerou, M, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-02-10 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2X48
 
 | | ORF 55 from Sulfolobus islandicus rudivirus 1 | | Descriptor: | CAG38821, PHOSPHATE ION | | Authors: | Oke, M, Carter, L, Johnson, K.A, Liu, H, Mcmahon, S, Naismith, J.H, White, M.F. | | Deposit date: | 2010-01-28 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2W8W
 
 | | N100Y SPT with PLP-ser | | Descriptor: | SERINE PALMITOYLTRANSFERASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2009-01-19 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The External-Aldimine Form of Serine Palmitoyltranserase; Structural, Kinetic and Spectroscopic Analysis of the Wild-Type Enzyme and Hsan1 Mutant Mimics.
J.Biol.Chem., 284, 2009
|
|
6ENK
 
 | | The X-ray crystal structure of DesE bound to desferrioxamine B | | Descriptor: | DesE, SODIUM ION, desferrioxamine B | | Authors: | Naismith, J.H, McMahon, S.A, Challis, G.L, Kadi, N, Oke, M, Liu, H, Carter, L.G, Johnson, K.A. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Desferrioxamine biosynthesis: diverse hydroxamate assembly by substrate-tolerant acyl transferase DesC.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
