7E30
 
 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E31
 
 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | Descriptor: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | Authors: | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E5J
 
 | | Crystal structure of beta-glucosidase from Thermoanaerobacterium saccharolyticum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase, SODIUM ION | | Authors: | Nam, K.H. | | Deposit date: | 2021-02-18 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Biochemical and Structural Analysis of a Glucose-Tolerant beta-Glucosidase from the Hemicellulose-Degrading Thermoanaerobacterium saccharolyticum.
Molecules, 27, 2022
|
|
4F60
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (T148L, G171Q, A172V, C176F). | | Descriptor: | FLUORIDE ION, Haloalkane dehalogenase | | Authors: | Plevaka, M, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4F5Z
 
 | | Crystal structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant (L95V, A172V). | | Descriptor: | BENZOIC ACID, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Kulik, D, Kuta-Smatanova, I, Rezacova, P. | | Deposit date: | 2012-05-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering enzyme stability and resistance to an organic cosolvent by modification of residues in the access tunnel.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
6S97
 
 | |
6S6E
 
 | |
3ZPH
 
 | |
3ZWQ
 
 | |
9FW1
 
 | |
9FVF
 
 | | UMG-SP3 amidase from uncultured bacterium | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Amidase UMG-SP3, SULFATE ION | | Authors: | Rotilio, L, Morth, J.P. | | Deposit date: | 2024-06-27 | | Release date: | 2025-02-05 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Characterization of an Amidase Targeting a Polyurethane for Sustainable Recycling.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9FZ1
 
 | |
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C9T
 
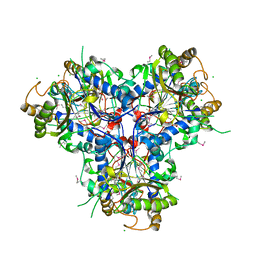 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7W69
 
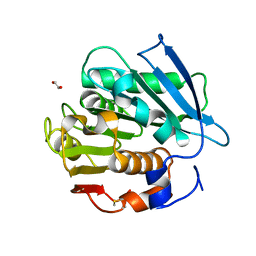 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W66
 
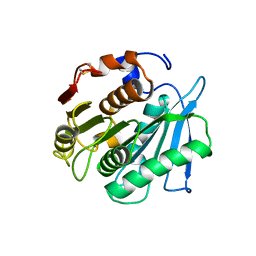 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
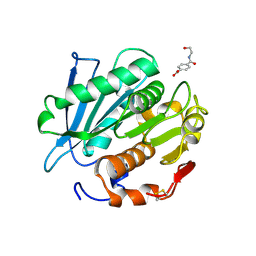 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W1I
 
 | | Crystal structure of carboxylesterase mutant from Thermobifida fusca with C8X and C9C | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, Carboxylesterase, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7W1J
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
7VPB
 
 | | Crystal structure of a novel hydrolase in apo form | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, ACETATE ION, plastic degrading hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural insight and engineering of a plastic degrading hydrolase Ple629.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
7W1K
 
 | | Crystal structure of carboxylesterase from Thermobifida fusca | | Descriptor: | Carboxylesterase | | Authors: | Han, X, Gerlis, H, Li, Z, Gao, J, Wei, R, Liu, W. | | Deposit date: | 2021-11-19 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Insights into (Tere)phthalate-Ester Hydrolysis by a Carboxylesterase and Its Role in Promoting PET Depolymerization
Acs Catalysis, 12, 2022
|
|
