2ZGR
 
 | |
2ZGS
 
 | |
3A0E
 
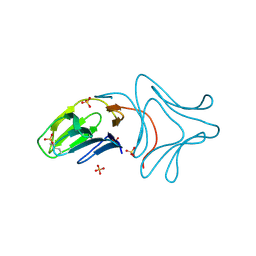 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
3A0C
 
 | |
3A0D
 
 | |
3AQS
 
 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AQT
 
 | | CRYSTAL STRUCTURE OF RolR (NCGL1110) complex WITH ligand RESORCINOL | | Descriptor: | Bacterial regulatory proteins, tetR family, RESORCINOL | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
3AYC
 
 | | Crystal structure of galectin-3 CRD domian complexed with GM1 pentasaccharide | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Galectin-3, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3AYE
 
 | |
3AYA
 
 | | Crystal structure of galectin-3 CRD domian complexed with Thomsen-Friedenreich antigen | | Descriptor: | Galectin-3, SULFATE ION, THREONINE, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3AYD
 
 | | Crystal structure of galectin-3 CRD domian complexed with TFN | | Descriptor: | Galectin-3, P-NITROPHENOL, SULFATE ION, ... | | Authors: | Bian, C.F, Li, D.F, Wang, D.C. | | Deposit date: | 2011-05-04 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for distinct binding properties of the human galectins to thomsen-friedenreich antigen
Plos One, 6, 2011
|
|
3NI2
 
 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
5HDK
 
 | |
5HDG
 
 | |
5HDN
 
 | |
5HSM
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 | | Descriptor: | SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5HSO
 
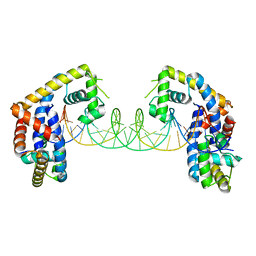 | | Crystal structure of MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN Rv2887 complex with DNA | | Descriptor: | DNA (30-MER), the upstream sequence of Rv0560c, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2016-01-26 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
1P9G
 
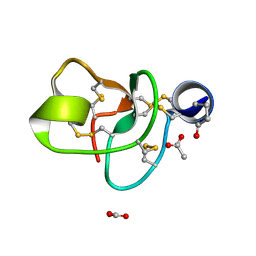 | | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Ecommia ulmoides Oliver at atomic resolution | | Descriptor: | ACETATE ION, EAFP 2 | | Authors: | Xiang, Y, Huang, R.H, Liu, X.Z, Wang, D.C. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution.
J.Struct.Biol., 148, 2004
|
|
5X80
 
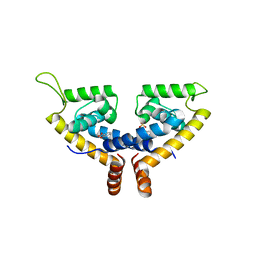 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 COMPLEX WITH SALICYLIC ACID | | Descriptor: | 2-HYDROXYBENZOIC ACID, SULFATE ION, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5X7Z
 
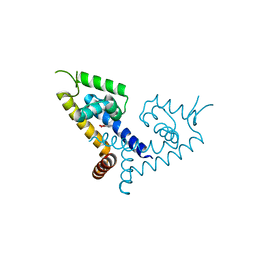 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS MARR FAMILY PROTEIN RV2887 COMPLEX WITH P-AMINOSALICYLIC ACID | | Descriptor: | 2-HYDROXY-4-AMINOBENZOIC ACID, Uncharacterized HTH-type transcriptional regulator Rv2887 | | Authors: | Gao, Y.R, Li, D.F, Wang, D.C, Bi, L.J. | | Deposit date: | 2017-02-28 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the regulatory mechanism of MarR protein Rv2887 in M. tuberculosis
Sci Rep, 7, 2017
|
|
5XUA
 
 | |
5XUB
 
 | |
5Y6G
 
 | | PilZ domain with c-di-GMP of YcgR from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
5Y6H
 
 | |
5Y6F
 
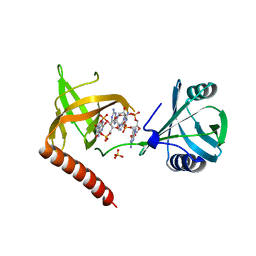 | | Crystal structure of YcgR in complex with c-di-GMP from Escherichia coli | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR, SULFATE ION | | Authors: | Hou, Y.J, Wang, D.C, Li, D.F. | | Deposit date: | 2017-08-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the mechanism of c-di-GMP-bound YcgR regulating flagellar motility inEscherichia coli.
J.Biol.Chem., 295, 2020
|
|
