2NMT
 
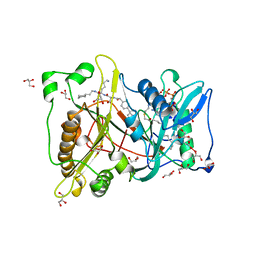 | | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE BOUND TO MYRISTOYL-COA AND PEPTIDE ANALOGS | | Descriptor: | GLYCEROL, MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, S-(2-OXO)PENTADECYLCOA, ... | | Authors: | Fuetterer, K, Bhatnagar, R.S, Waksman, G. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of N-myristoyltransferase with bound myristoylCoA and peptide substrate analogs.
Nat.Struct.Biol., 5, 1998
|
|
2OFQ
 
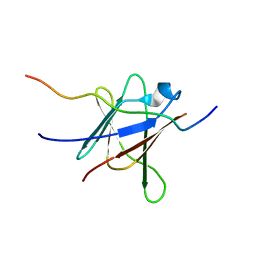 | | NMR Solution Structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system | | Descriptor: | TraN, TraO | | Authors: | Harris, R, Bayliss, R, Driscoll, P.C, Waksman, G. | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PT7
 
 | | Crystal structure of Cag VirB11 (HP0525) and an inhibitory protein (HP1451) | | Descriptor: | Cag-alfa, Hypothetical protein | | Authors: | Hare, S, Fischer, W, Williams, R, Terradot, L, Bayliss, R, Haas, R, Waksman, G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase.
Embo J., 26, 2007
|
|
3U4K
 
 | |
3CRF
 
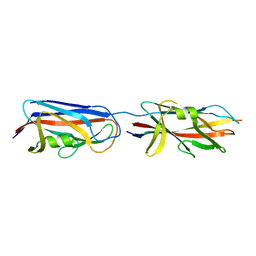 | |
3JQO
 
 | | Crystal structure of the outer membrane complex of a type IV secretion system | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LAURYL DIMETHYLAMINE-N-OXIDE, TraF protein, ... | | Authors: | Chandran, V, Fronzes, R, Duquerroy, S, Cronin, N, Navaza, J, Waksman, G. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the outer membrane complex of a type IV secretion system
Nature, 462, 2009
|
|
3KTQ
 
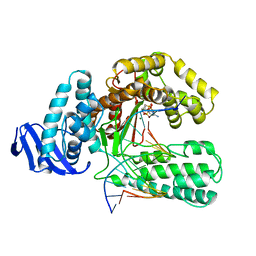 | |
3LFU
 
 | | Crystal Structure of E. coli UvrD | | Descriptor: | DNA helicase II, SULFATE ION | | Authors: | Korolev, S, Waksman, G, Lohman, T.M. | | Deposit date: | 2010-01-18 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding.
J.Mol.Biol., 411, 2011
|
|
4AG6
 
 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | SULFATE ION, TYPE IV SECRETORY PATHWAY VIRB4 COMPONENTS-LIKE PROTEIN | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AG5
 
 | | Structure of VirB4 of Thermoanaerobacter pseudethanolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wallden, K, Williams, R, Yan, J, Lian, P.W, Wang, L, Thalassinos, K, Orlova, E.V, Waksman, G. | | Deposit date: | 2012-01-24 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Virb4 ATPase, Alone and Bound to the Core Complex of a Type Iv Secretion System.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZBJ
 
 | | Fitting results in the I-layer of the subnanometer structure of the bacterial pKM101 type IV secretion system core complex digested with elastase | | Descriptor: | TRAO PROTEIN | | Authors: | Rivera-Calzada, A, Fronzes, R, Savva, C.G, Chandran, V, Lian, P.W, Laeremans, T, Pardon, E, Steyaert, J, Remaut, H, Waksman, G, Orlova, E.V. | | Deposit date: | 2012-11-10 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Structure of a Bacterial Type Iv Secretion Core Complex at Subnanometre Resolution.
Embo J., 32, 2013
|
|
2GR7
 
 | | Hia 992-1098 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Adhesin | | Authors: | Meng, G, Waksman, G. | | Deposit date: | 2006-04-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter.
Embo J., 25, 2006
|
|
2W07
 
 | | Structural determinants of polymerization reactivity of the P pilus adaptor subunit PapF | | Descriptor: | CHAPERONE PROTEIN PAPD, MINOR PILIN SUBUNIT PAPF, SULFATE ION | | Authors: | Verger, D, Rose, R.J, Paci, E, Costakes, G, Daviter, T, Hultgren, S, Remaut, H, Ashcroft, A.E, Radford, S.E, Waksman, G. | | Deposit date: | 2008-08-12 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants of Polymerization Reactivity of the P Pilus Adaptor Subunit Papf.
Structure, 16, 2008
|
|
2XG5
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 5d | | Descriptor: | (2R)-2-[5-CYCLOPROPYL-6-(HYDROXYSULFANYL)-4-(NAPHTHALEN-1-YLMETHYL)-2-OXOPYRIDIN-1(2H)-YL]-3-PHENYLPROPANOIC ACID, (2R,3R)-8-CYCLOPROPYL-7-(NAPHTHALEN-1-YLMETHYL)-5-OXO-2-PHENYL-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
2XG4
 
 | | E. coli P pilus chaperone-subunit complex PapD-PapH bound to pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD, COBALT (II) ION, ... | | Authors: | Remaut, H, Phan, G, Buelens, F, Chorell, E, Pinkner, J.S, Edvinsson, S, Almqvist, F, Hultgren, S.J, Waksman, G. | | Deposit date: | 2010-05-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Synthesis of C-2 Substituted Thiazolo and Dihydrothiazolo Ring-Fused 2-Pyridones: Pilicides with Increased Antivirulence Activity.
J.Med.Chem., 53, 2010
|
|
3EMO
 
 | |
3EMI
 
 | |
3EMF
 
 | |
2GR8
 
 | | Hia 1022-1098 | | Descriptor: | Adhesin | | Authors: | Meng, G, Waksman, G. | | Deposit date: | 2006-04-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter.
Embo J., 25, 2006
|
|
5N8O
 
 | |
1KA8
 
 | | Crystal Structure of the Phage P4 Origin-Binding Domain | | Descriptor: | putative P4-specific DNA primase | | Authors: | Yeo, H.J, Ziegelin, G, Korolev, S, Calendar, R, Lanka, E, Waksman, G. | | Deposit date: | 2001-10-31 | | Release date: | 2002-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Phage P4 origin-binding domain structure reveals a mechanism for regulation of DNA-binding activity by homo- and heterodimerization of winged helix proteins.
Mol.Microbiol., 43, 2002
|
|
7OVB
 
 | | L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains | | Descriptor: | DotY, DotZ, IcmJ (DotN), ... | | Authors: | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proteins DotY and DotZ modulate the dynamics and localization of the type IVB coupling complex of Legionella pneumophila.
Mol.Microbiol., 117, 2022
|
|
1J8S
 
 | | PAPG ADHESIN RECEPTOR BINDING DOMAIN-UNBOUND FORM | | Descriptor: | PYELONEPHRITIC ADHESIN | | Authors: | Dodson, K.W, Pinkner, J.S, Rose, T, Magnusson, G, Hultgren, S.J, Waksman, G. | | Deposit date: | 2001-05-22 | | Release date: | 2001-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor.
Cell(Cambridge,Mass.), 105, 2001
|
|
5OH0
 
 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-22 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
2J7L
 
 | | E. coli P Pilus chaperone PapD in complex with a pilus biogenesis inhibitor, pilicide 2c | | Descriptor: | (3R)-8-CYCLOPROPYL-6-(MORPHOLIN-4-YLMETHYL)-7-(1-NAPHTHYLMETHYL)-5-OXO-2,3-DIHYDRO-5H-[1,3]THIAZOLO[3,2-A]PYRIDINE-3-CARBOXYLIC ACID, CHAPERONE PROTEIN PAPD | | Authors: | Remaut, H, Pinkner, J.S, Hultgren, S.J, Almqvist, F, Waksman, G. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rationally Designed Small Compounds Inhibit Pilus Biogenesis in Uropathogenic Bacteria.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
