7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
3WY9
 
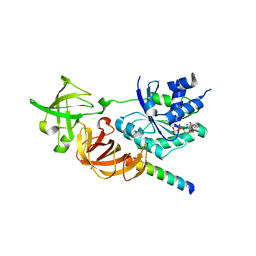 | | Crystal structure of a complex of the archaeal ribosomal stalk protein aP1 and the GDP-bound archaeal elongation factor aEF1alpha | | Descriptor: | 50S ribosomal protein L12, Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
5EO8
 
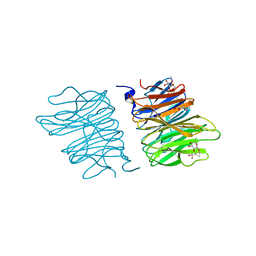 | | Crystal structure of AOL(868) | | Descriptor: | Predicted protein, methyl 1-seleno-beta-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5EO7
 
 | | Crystal structure of AOL | | Descriptor: | Predicted protein, SULFATE ION, methyl 1-seleno-alpha-L-fucopyranoside | | Authors: | Kato, R, Kiso, M, Ishida, H, Ando, H, Suzuki, T, Shimabukuro, S, Makyio, H. | | Deposit date: | 2015-11-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Six independent fucose-binding sites in the crystal structure of Aspergillus oryzae lectin
Biochem.Biophys.Res.Commun., 477, 2016
|
|
3WYA
 
 | | Crystal structure of GDP-bound EF1alpha from Pyrococcus horikoshii | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ito, K, Honda, T, Suzuki, T, Miyoshi, T, Murakami, R, Yao, M, Uchiumi, T. | | Deposit date: | 2014-08-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into the interaction of the ribosomal stalk protein with elongation factor 1 alpha.
Nucleic Acids Res., 42, 2014
|
|
1WTG
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-biphenylalanine-Gln-p-aminobenzamidine | | Descriptor: | 2-(3-BIPHENYL-4-YL-2-ETHANESULFONYLAMINO-PROPIONYLAMINO)-PENTANEDIOIC ACID 5-AMIDE 1-(4-CARBAMIMIDOYL-BENZYLAMIDE), CALCIUM ION, Coagulation factor VII, ... | | Authors: | Kadono, S, Sakamoto, S, Kikuchi, Y, Oh-Eda, M, Yabuta, N, Kitazawa, K, Yoshihashi, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Kodama, M, Haramura, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-11-23 | | Release date: | 2005-11-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Novel interactions of large P3 moiety and small P4 moiety in the binding of the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 326, 2005
|
|
7WEM
 
 | | Solid-state NMR Structure of TFo c-Subunit Ring | | Descriptor: | ATP synthase subunit c | | Authors: | Akutsu, H, Todokoro, Y, Kang, S.-J, Suzuki, T, Yoshida, M, Ikegami, T, Fujiwara, T. | | Deposit date: | 2021-12-23 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Chemical Conformation of the Essential Glutamate Site of the c -Ring within Thermophilic Bacillus F o F 1 -ATP Synthase Determined by Solid-State NMR Based on its Isolated c -Ring Structure.
J.Am.Chem.Soc., 144, 2022
|
|
1WUN
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-Trp-Gln-p-aminobenzamidine | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-(ETHYLSULFONYL)TRYPTOPHYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}GLUTAMAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Yoshihashi, K, Kitazawa, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-12-08 | | Release date: | 2005-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 327, 2005
|
|
2UXC
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA UCGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACGA, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
1WV7
 
 | | Human Factor Viia-Tissue Factor Complexed with ethylsulfonamide-D-5-propoxy-Trp-Gln-p-aminobenzamidine | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-(ETHYLSULFONYL)-5-PROPOXY-L-TRYPTOPHYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}-L-GLUTAMAMIDE, ... | | Authors: | Kadono, S, Sakamoto, A, Kikuchi, Y, Oh-eda, M, Yabuta, N, Yoshihashi, K, Kitazawa, T, Suzuki, T, Koga, T, Hattori, K, Shiraishi, T, Haramura, M, Kodama, H, Ono, Y, Esaki, T, Sato, H, Watanabe, Y, Itoh, S, Ohta, M, Kozono, T. | | Deposit date: | 2004-12-11 | | Release date: | 2005-12-11 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor
Biochem.Biophys.Res.Commun., 327, 2005
|
|
2UXD
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA CGGG in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT CGGG, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON CCCG, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Trnas with an Expanded Anticodon Loop in the Decoding Center of the 30S Ribosomal Subunit.
RNA, 13, 2007
|
|
2UXB
 
 | | Crystal structure of an extended tRNA anticodon stem loop in complex with its cognate mRNA GGGU in the context of the Thermus thermophilus 30S subunit. | | Descriptor: | 16S RIBOSOMAL RNA, A-SITE MESSENGER RNA FRAGMENT GGGU, ANTICODON STEM-LOOP OF TRANSFER RNA WITH ANTICODON ACCC, ... | | Authors: | Dunham, C.M, Selmer, M, Phelps, S.S, Kelley, A.C, Suzuki, T, Joseph, S, Ramakrishnan, V. | | Deposit date: | 2007-03-28 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit.
RNA, 13, 2007
|
|
4XD7
 
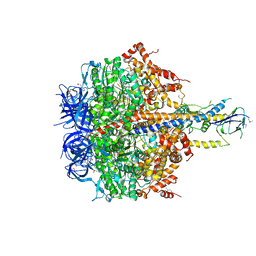 | | Structure of thermophilic F1-ATPase inhibited by epsilon subunit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | SHIRAKIHARA, Y, SHIRATORI, A, TANIKAWA, H, NAKASAKO, M, YOSHIDA, M, SUZUKI, T. | | Deposit date: | 2014-12-19 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure of a thermophilic F1 -ATPase inhibited by an epsilon-subunit: deeper insight into the epsilon-inhibition mechanism.
Febs J., 282, 2015
|
|
7VNW
 
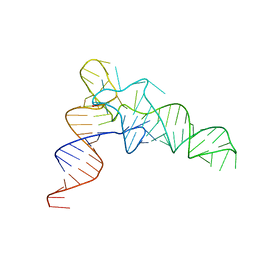 | |
7VNV
 
 | |
7VNX
 
 | | Crystal structure of TkArkI | | Descriptor: | GUANOSINE, TkArkI | | Authors: | Yamashita, S, Minowa, K, Ohira, T, Suzuki, T, Tomita, K. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Reversible RNA phosphorylation stabilizes tRNA for cellular thermotolerance.
Nature, 605, 2022
|
|
5B2H
 
 | | Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi | | Descriptor: | HA-33, TRIETHYLENE GLYCOL | | Authors: | Akiyama, T, Hayashi, S, Matsumoto, T, Hasegawa, K, Yamano, A, Suzuki, T, Niwa, K, Watanabe, T, Sagane, Y, Yajima, S. | | Deposit date: | 2016-01-15 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational divergence in the HA-33/HA-17 trimer of serotype C and D botulinum toxin complex
Biochem.Biophys.Res.Commun., 476, 2016
|
|
1WU0
 
 | |
1WPW
 
 | | Crystal Structure of IPMDH from Sulfolobus tokodaii | | Descriptor: | 3-isopropylmalate dehydrogenase, MAGNESIUM ION | | Authors: | Hirose, R, Sakurai, M, Suzuki, T, Moriyama, H, Sato, T, Yamagishi, A, Oshima, T, Tanaka, N. | | Deposit date: | 2004-09-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of IPMDH from Sulfolobus tokodaii
To be Published
|
|
5ZR3
 
 | | Crystal structure of Hsp90-alpha N-terminal domain in complex with 4-(3-isopropyl-4-(4-(1-methyl-1H-pyrazol-4-yl)-1H-imidazol-1-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl)-3-methylbenzamide | | Descriptor: | 3-methyl-4-{4-[4-(1-methyl-1H-pyrazol-4-yl)-1H-imidazol-1-yl]-3-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-1-yl}benzamide, Heat shock protein HSP 90-alpha | | Authors: | Uno, T, Chong, K.T, Suzuki, T. | | Deposit date: | 2018-04-23 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 3-Ethyl-4-(3-isopropyl-4-(4-(1-methyl-1 H-pyrazol-4-yl)-1 H-imidazol-1-yl)-1 H-pyrazolo[3,4- b]pyridin-1-yl)benzamide (TAS-116) as a Potent, Selective, and Orally Available HSP90 Inhibitor.
J. Med. Chem., 62, 2019
|
|
8YUQ
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and dA4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUP
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and A4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-28 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUR
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and Am4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUS
 
 | | E. coli 70S ribosome complexed with P.putida tRNAIle2 and A(F)4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
