6LWQ
 
 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing a C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*TP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
6LWR
 
 | | Crystal structure of human NEIL1(K242) bound to duplex DNA containing a cleaved C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*(PDA))-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
5I0A
 
 | | RecA mini intein in complex with cisplatin | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, Cisplatin, Intein, ... | | Authors: | Li, Z, Zhang, J, Li, H.M. | | Deposit date: | 2016-02-03 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into protein splicing inhibition by platinum therapeutics as potential anti-microbials
To be published
|
|
7XJF
 
 | | Crystal structure of 6MW3211 Fab in complex with CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2022-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Theranostics, 13, 2023
|
|
6VXN
 
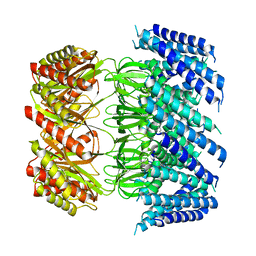 | | Cryo-EM structure of Arabidopsis thaliana MSL1 A320V | | Descriptor: | DODECANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
5GO0
 
 | |
3TKZ
 
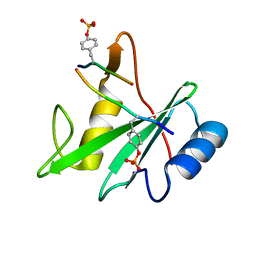 | | Structure of the SHP-2 N-SH2 domain in a 1:2 complex with RVIpYFVPLNR peptide | | Descriptor: | PROTEIN (RVIpYFVPLNR peptide), Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-10-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
3TL0
 
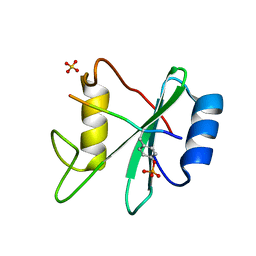 | | Structure of SHP2 N-SH2 domain in complex with RLNpYAQLWHR peptide | | Descriptor: | RLNpYAQLWHR peptide, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Y, Zhang, J, Yuan, C, Hard, R.L, Park, I.H, Li, C, Bell, C.E, Pei, D. | | Deposit date: | 2011-08-29 | | Release date: | 2011-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Simultaneous binding of two peptidyl ligands by a SRC homology 2 domain.
Biochemistry, 50, 2011
|
|
8H4Q
 
 | |
8H4H
 
 | |
9C9R
 
 | |
9C9Q
 
 | |
6VXM
 
 | | Cryo-EM structure of Arabidopsis thaliana MSL1 | | Descriptor: | EICOSANE, Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Deng, Z, Zhang, J, Yuan, P. | | Deposit date: | 2020-02-22 | | Release date: | 2020-08-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural mechanism for gating of a eukaryotic mechanosensitive channel of small conductance.
Nat Commun, 11, 2020
|
|
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
6VXP
 
 | |
5HNK
 
 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HRB
 
 | | The crystal structure of AsfvPolX:DNA1 binary complex | | Descriptor: | BETA-MERCAPTOETHANOL, DNA (5'-D(*CP*GP*GP*AP*TP*AP*TP*CP*C)-3'), DNA polymerase beta-like protein, ... | | Authors: | Chen, Y.Q, Zhang, J, Gan, J.H. | | Deposit date: | 2016-01-23 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of Se-AsfvPolX(L52/163M mutant) in complex with 1nt-gap DNA1
To Be Published
|
|
5HRH
 
 | |
6K8J
 
 | | Structure of NLRC4 CARD filament | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Gong, Q, Xu, C, Zhang, J, Boo, Z.Z, Wu, B. | | Deposit date: | 2019-06-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for distinct inflammasome complex assembly by human NLRP1 and CARD8.
Nat Commun, 12, 2021
|
|
2M1H
 
 | | Solution structure of a PWWP domain from Trypanosoma brucei | | Descriptor: | Transcription elongation factor S-II | | Authors: | Wang, R, Fan, K, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-11-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of TbTFIIS2-1 PWWP domain from Trypanosoma brucei.
Proteins, 84, 2016
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
4LYC
 
 | | Cd ions within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
8WZP
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with CCF0058981 | | Descriptor: | 2-(benzotriazol-1-yl)-~{N}-[(3-chlorophenyl)methyl]-~{N}-[4-(1~{H}-imidazol-5-yl)phenyl]ethanamide, 3C-like proteinase nsp5 | | Authors: | Jiang, H.H, Zou, X.F, Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease (M pro ) mutants in complex with the non-covalent inhibitor CCF0058981.
Biochem.Biophys.Res.Commun., 692, 2024
|
|
4LYB
 
 | | CdS within a lysoyzme single crystal | | Descriptor: | CADMIUM ION, Lysozyme C | | Authors: | Wei, H, House, S, Wu, J, Zhang, J, Wang, Z, He, Y, Gao, Y.-G, Robinson, H, Li, W, Zuo, J.-M, Robertson, I.M, Lu, Y. | | Deposit date: | 2013-07-30 | | Release date: | 2015-02-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Enhanced and tunable fluorescent quantum dots within a single crystal of protein
TO BE PUBLISHED
|
|
7CA8
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Li, J, Zhang, J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode.
Sci Bull (Beijing), 66, 2021
|
|
