2YVQ
 
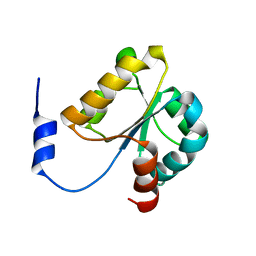 | | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens | | Descriptor: | Carbamoyl-phosphate synthase | | Authors: | Xie, Y, Ihsanawati, Kishishita, S, Murayama, K, Takemoto, C, Shirozu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of MGS domain of carbamoyl-phosphate synthetase from homo sapiens
To be Published
|
|
2YY9
 
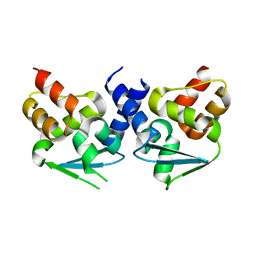 | | Crystal structure of BTB domain from mouse HKR3 | | Descriptor: | Zinc finger and BTB domain-containing protein 48 | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of BTB domain from mouse HKR3
To be Published
|
|
2YZ2
 
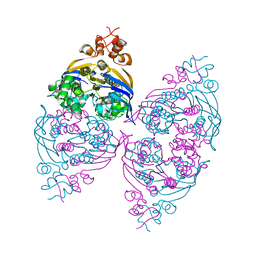 | | Crystal structure of the ABC transporter in the cobalt transport system | | Descriptor: | Putative ABC transporter ATP-binding protein TM_0222 | | Authors: | Ethayathullah, A.S, Bessho, Y, Padmanabhan, B, Singh, T.P, Kaur, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-02 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the ABC transporter in the cobalt transport system
To be Published
|
|
2YZU
 
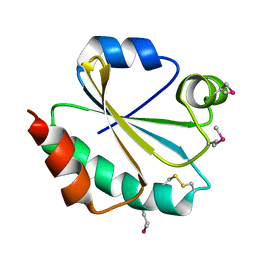 | |
2Z02
 
 | | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase wit ATP from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, Phosphoribosylaminoimidazole-succinocarboxamide synthase, ... | | Authors: | Kanagawa, M, Baba, S, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of phosphoribosylaminoimidazolesuccinocarboxamide synthase from Methanocaldococcus jannaschii
To be Published
|
|
2YQF
 
 | | Solution structure of the death domain of Ankyrin-1 | | Descriptor: | Ankyrin-1 | | Authors: | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the death domain of Ankyrin-1
To be Published
|
|
2YRJ
 
 | | Solution structure of the C2H2-type zinc finger domain (781-813) from zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Qin, X.R, Izumi, K, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2-type zinc finger domain (781-813) from zinc finger protein 473
To be Published
|
|
2YRZ
 
 | | Solution structure of the fibronectin type III domain of human Integrin beta-4 | | Descriptor: | Integrin beta-4 | | Authors: | Abe, H, Sasagawa, A, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fibronectin type III domain of human Integrin beta-4
To be Published
|
|
2YSH
 
 | | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7 | | Descriptor: | Growth-arrest-specific protein 7 | | Authors: | Ohnishi, S, Tochio, N, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the WW domain from the human growth-arrest-specific protein 7, GAS-7
To be Published
|
|
2YSX
 
 | | Solution structure of the human SHIP SH2 domain | | Descriptor: | Signaling inositol polyphosphate phosphatase SHIP II | | Authors: | Kasai, T, Koshiba, S, Watanabe, S, Harada, T, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the human SHIP SH2 domain
To be Published
|
|
2YTJ
 
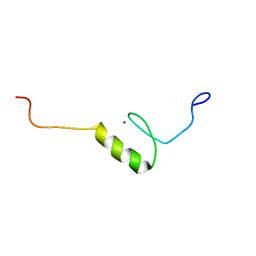 | | Solution structure of the C2H2 type zinc finger (region 771-803) of human Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Tomizawa, T, Tochio, N, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 771-803) of human Zinc finger protein 484
To be Published
|
|
2YR3
 
 | | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle | | Descriptor: | Myosin light chain kinase, smooth muscle | | Authors: | Qin, X.R, Kurosaki, C, Yoshida, M, Hayahsi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth Ig-like domain from myosin light chain kinase, smooth muscle
To be Published
|
|
2YRV
 
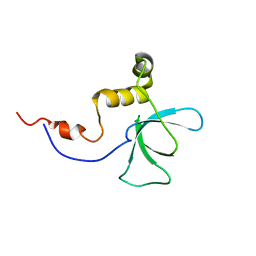 | | Solution structure of the RBB1NT domain of human RB(retinoblastoma)-binding protein 1 | | Descriptor: | AT-rich interactive domain-containing protein 4A | | Authors: | Nameki, N, Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RBB1NT domain of human RB(retinoblastoma)-binding protein 1
To be Published
|
|
2YSF
 
 | | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH | | Descriptor: | E3 ubiquitin-protein ligase Itchy homolog | | Authors: | Ohnishi, S, Li, H, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fourth WW domain from the human E3 ubiquitin-protein ligase Itchy homolog, ITCH
To be Published
|
|
2YSV
 
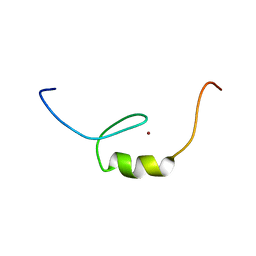 | | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tsuda, K, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-04 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of C2H2 type Zinc finger domain 17 in Zinc finger protein 473
To be Published
|
|
2YUJ
 
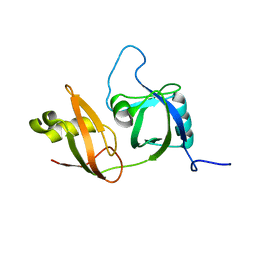 | |
2YWQ
 
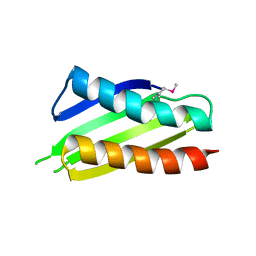 | | Crystal structure of Thermus thermophilus Protein Y N-terminal domain | | Descriptor: | Ribosomal subunit interface protein | | Authors: | Kawazoe, M, Takemoto, C, Kaminishi, T, Tatsuguchi, A, Saito, Y, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-21 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of Thermus thermophilus Protein Y N-terminal domain
To be Published
|
|
2YX8
 
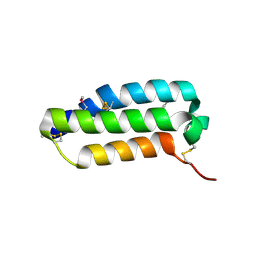 | | Crystal structure of the extracellular domain of human RAMP1 | | Descriptor: | Receptor activity-modifying protein 1 | | Authors: | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Shindo, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human receptor activity-modifying protein 1 extracellular domain.
Protein Sci., 17, 2008
|
|
2YTM
 
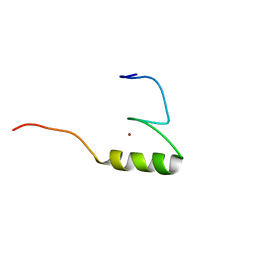 | | Solution structure of the C2H2 type zinc finger (region 696-728) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 696-728) of human Zinc finger protein 28 homolog
To be Published
|
|
2YUC
 
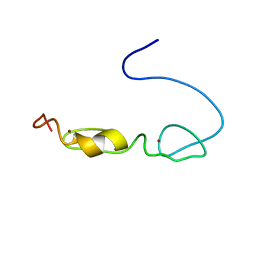 | |
2YUN
 
 | | Solution structure of the SH3 domain of human Nostrin | | Descriptor: | Nostrin | | Authors: | Abe, H, Tochio, N, Miyamoto, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human Nostrin
To be Published
|
|
2YUT
 
 | |
2YZH
 
 | | Crystal structure of peroxiredoxin-like protein from Aquifex aeolicus | | Descriptor: | Probable thiol peroxidase, SULFATE ION | | Authors: | Ebihara, A, Manzoku, M, Fujimoto, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peroxiredoxin-like protein from Aquifex aeolicus
to be published
|
|
2Z0A
 
 | | Crystal structure of RNA-binding domain of NS1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1) | | Descriptor: | GLYCINE, Nonstructural protein 1, SUCCINIC ACID | | Authors: | Saijo, S, Kishishita, S, Kamo-Uchikubo, T, Terada, T, Shirouzu, M, Ito, H, Ito, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of RNA-binding domain of NS1 from influenza A virus A/crow/Kyoto/T1/2004(H5N1)
To be Published
|
|
2YUZ
 
 | | Solution Structure of 4th Immunoglobulin Domain of Slow Type Myosin-Binding Protein C | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Niraula, T.N, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-06 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 4th Immunoglobulin Domain of Slow Type Myosin-Binding Protein C
To be Published
|
|
