6DQE
 
 | |
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXX
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 6 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
6DQD
 
 | |
8CXS
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DNA Strand 1, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXV
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 3 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXY
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(2-Phenethyl)adenosine (Compound 8) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXU
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor APNEA (Compound 9) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXT
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-benzyladenosine (Compound 1) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXW
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor piclidenoson (Compound 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY0
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor MC4756 (Compound 178) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(4-phenylbutyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CXZ
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor N6-(3-Phenylpropyl)adenosine (Compound 14) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(3-phenylpropyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
6DQ7
 
 | |
6DQC
 
 | |
1ML9
 
 | | Structure of the Neurospora SET domain protein DIM-5, a histone lysine methyltransferase | | Descriptor: | Histone H3 methyltransferase DIM-5, UNKNOWN LIGAND, ZINC ION | | Authors: | Zhang, X, Tamaru, H, Khan, S.I, Horton, J.R, Keefe, L.J, Selker, E.U, Cheng, X. | | Deposit date: | 2002-08-30 | | Release date: | 2002-10-23 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Neurospora SET domain protein DIM-5,
a histone H3 lysine methyltransferase
Cell(Cambridge,Mass.), 111, 2002
|
|
1MR3
 
 | | Saccharomyces cerevisiae ADP-ribosylation Factor 2 (ScArf2) complexed with GDP-3'P at 1.6A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ADP-ribosylation factor 2, ... | | Authors: | Amor, J.-C, Horton, J.R, Zhu, X, Wang, Y, Sullards, C, Ringe, D, Cheng, X, Kahn, R.A. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases.
J.Biol.Chem., 276, 2001
|
|
1NGN
 
 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
3EU9
 
 | | The ankyrin repeat domain of Huntingtin interacting protein 14 | | Descriptor: | GLYCEROL, HISTIDINE, Huntingtin-interacting protein 14, ... | | Authors: | Gao, T, Collins, R.E, Horton, J.R, Zhang, R, Zhang, X, Cheng, X. | | Deposit date: | 2008-10-09 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The ankyrin repeat domain of Huntingtin interacting protein 14 contains a surface aromatic cage, a potential site for methyl-lysine binding.
Proteins, 76, 2009
|
|
3FPD
 
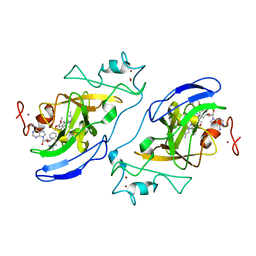 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1ORI
 
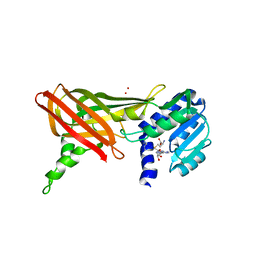 | | Structure of the predominant protein arginine methyltransferase PRMT1 | | Descriptor: | Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN LIGAND | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of its Binding to Substrate Peptides
Structure, 11, 2003
|
|
1OR8
 
 | | Structure of the Predominant protein arginine methyltransferase PRMT1 | | Descriptor: | GLYCEROL, Protein arginine N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, X, Cheng, X. | | Deposit date: | 2003-03-12 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Predominant Protein Arginine Methyltransferase PRMT1 and Analysis of Its Binding to Substrate Peptides
Structure, 11, 2003
|
|
