3AX6
 
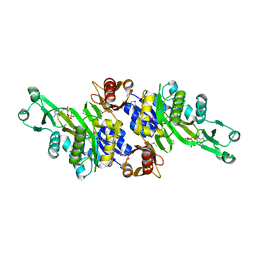 | | Crystal structure of N5-carboxyaminoimidazole ribonucleotide synthetase from Thermotoga maritima | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoribosylaminoimidazole carboxylase, ATPase subunit | | Authors: | Miyazawa, R, Kanagawa, M, Baba, S, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-03-30 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
To be Published
|
|
3AXT
 
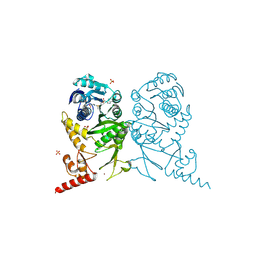 | | Complex structure of tRNA methyltransferase Trm1 from Aquifex aeolicus with S-adenosyl-L-Methionine | | Descriptor: | Probable N(2),N(2)-dimethylguanosine tRNA methyltransferase Trm1, S-ADENOSYLMETHIONINE, SULFATE ION, ... | | Authors: | Ihsanawati, Sengoku, T, Yokoyama, S, Bessho, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-04-14 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure
J.Biol.Chem., 286, 2011
|
|
3CM0
 
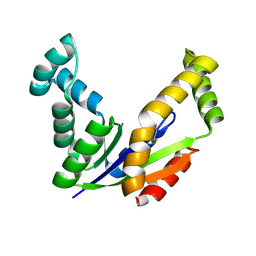 | | Crystal structure of adenylate kinase from Thermus thermophilus HB8 | | Descriptor: | Adenylate kinase | | Authors: | Nakagawa, N, Kondo, N, Masui, R, Matsuda, Z, Iwamoto, I, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-03-20 | | Release date: | 2008-09-23 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of adenylate kinase from Thermus thermophilus HB8.
To be Published
|
|
1V5N
 
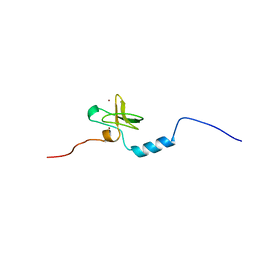 | | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana | | Descriptor: | PDI-like Hypothetical Protein At1g60420, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of DC1 Domain of PDI-like Hypothetical Protein from Arabidopsis thaliana
To be Published
|
|
2ZIN
 
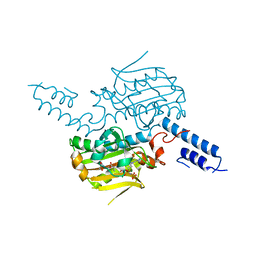 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and an ATP analogue | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, ... | | Authors: | Yanagisawa, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-02-19 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode N(varepsilon)-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification
Chem.Biol., 15, 2008
|
|
2ZKT
 
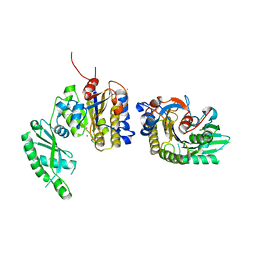 | |
1UJO
 
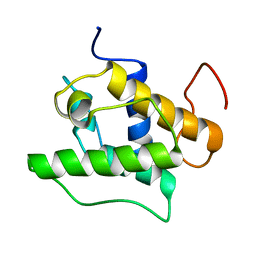 | |
1V65
 
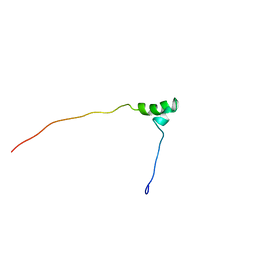 | | Solution structure of the Kruppel-associated box (KRAB) domain | | Descriptor: | RIKEN cDNA 2610044O15 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-27 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Kruppel-associated box (KRAB) domain
To be Published
|
|
1UL7
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
1V5K
 
 | | Solution structure of the CH domain from mouse EB-1 | | Descriptor: | microtubule-associated protein, RP/EB family, member 1 | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-25 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from mouse EB-1
To be Published
|
|
1WHH
 
 | | Solution structure of the 2nd CAP-Gly domain in mouse CLIP170-related 59kDa protein CLIPR-59 | | Descriptor: | CLIPR-59 | | Authors: | Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 2nd CAP-Gly domain in mouse CLIP170-related 59kDa protein CLIPR-59
To be Published
|
|
1WLV
 
 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1X2R
 
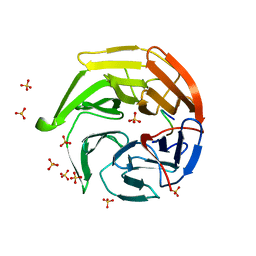 | | Structural basis for the defects of human lung cancer somatic mutations in the repression activity of Keap1 on Nrf2 | | Descriptor: | Kelch-like ECH-associated protein 1, Nuclear factor erythroid 2 related factor 2, SULFATE ION | | Authors: | Padmanabhan, B, Tong, K.I, Nakamura, Y, Ohta, T, Scharlock, M, Kobayashi, A, Ohtsuji, M, Kang, M.-I, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-04-26 | | Release date: | 2006-03-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for defects of keap1 activity provoked by its point mutations in lung cancer
Mol.Cell, 21, 2006
|
|
1WM6
 
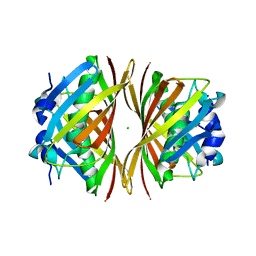 | |
1WLU
 
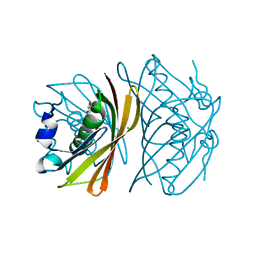 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, phenylacetic acid degradation protein PaaI | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-06-29 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1WN3
 
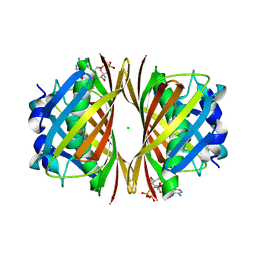 | | Crystal structure of TT0310 protein from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, CHLORIDE ION, HEXANOYL-COENZYME A, ... | | Authors: | Kunishima, N, Sugahara, M, Miyano, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Novel Induced-fit Reaction Mechanism of Asymmetric Hot Dog Thioesterase PaaI
J.Mol.Biol., 352, 2005
|
|
1X8X
 
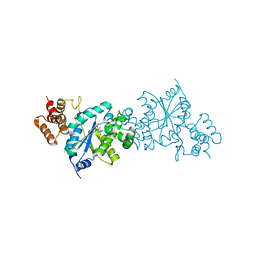 | | Tyrosyl t-RNA Synthetase from E.coli Complexed with Tyrosine | | Descriptor: | SULFATE ION, TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Takimura, T, Sekine, R, Kelly, V.P, Kamata, K, Sakamoto, K, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-08-19 | | Release date: | 2005-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Snapshots of the KMSKS Loop Rearrangement for Amino Acid Activation by Bacterial Tyrosyl-tRNA Synthetase
J.MOL.BIOL., 346, 2005
|
|
1VFG
 
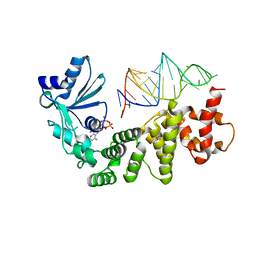 | | Crystal structure of tRNA nucleotidyltransferase complexed with a primer tRNA and an incoming ATP analog | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA (75-MER), poly A polymerase | | Authors: | Tomita, K, Fukai, S, Ishitani, R, Ueda, T, Takeuchi, N, Vassylyev, D.G, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for template-independent RNA polymerization.
Nature, 430, 2004
|
|
3AQA
 
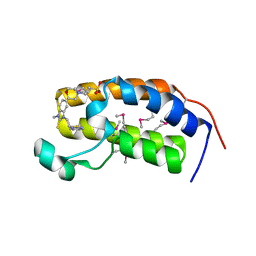 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
2YQZ
 
 | |
2YRC
 
 | |
2YRN
 
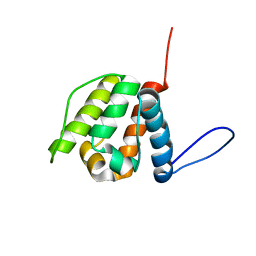 | | Solution structure of the CH domain from Human Neuron navigator 2 | | Descriptor: | Neuron navigator 2 isoform 4 | | Authors: | Tomizawa, T, Tochio, N, Koshiba, S, Inoue, M, Nakamura, Y, Furukawa, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CH domain from Human Neuron navigator 2
To be Published
|
|
2YSA
 
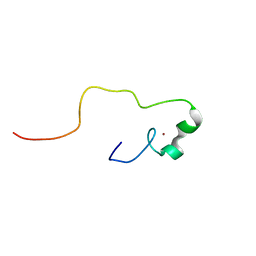 | | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1) | | Descriptor: | Retinoblastoma-binding protein 6, ZINC ION | | Authors: | Ohnishi, S, Sato, M, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the zinc finger CCHC domain from the human retinoblastoma-binding protein 6 (Retinoblastoma-binding Q protein 1, RBQ-1)
To be Published
|
|
2YQG
 
 | |
2YRW
 
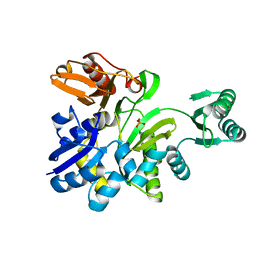 | | Crystal structure of GAR synthetase from Geobacillus kaustophilus | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide synthetase | | Authors: | Baba, S, Kanagawa, M, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
