4MJ6
 
 | | Crystal Structure of HLA-A*1101 in complex with H7-22, an influenza A(H7N9) virus epitope | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Liu, J, Tan, S, Zhao, M, Qi, J, Gao, G.F. | | Deposit date: | 2013-09-03 | | Release date: | 2014-10-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Cross-immunity Against Avian Influenza A(H7N9) Virus in the Healthy Population Is Affected by Antigenicity-Dependent Substitutions.
J.Infect.Dis., 214, 2016
|
|
5I77
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
6M2Z
 
 | | Crystal structure of a formolase, BFD variant M3 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
6M2Y
 
 | | Crystal structure of a formolase, BFD variant M6 from Pseudomonas putida | | Descriptor: | Benzoylformate decarboxylase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Wei, H.L, Liu, W.D, Li, T.Z, Zhu, L.L. | | Deposit date: | 2020-03-02 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Totally atom-economical synthesis of lactic acid from formaldehyde: combined bio-carboligation and chemo-rearrangement without the isolation of intermediate.
Green Chem, 22, 2020
|
|
8HE4
 
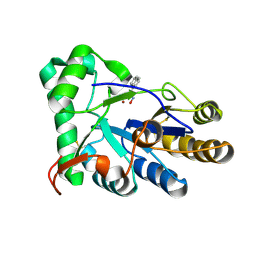 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, ~{N}-oxidanylnaphthalene-1-carboxamide | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE2
 
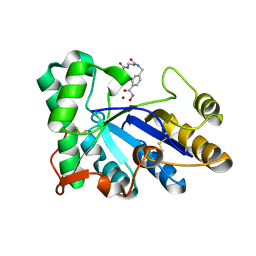 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION, tert-butyl N-[3-[[4-(oxidanylcarbamoyl)phenyl]methylamino]-3-oxidanylidene-propyl]carbamate | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HE1
 
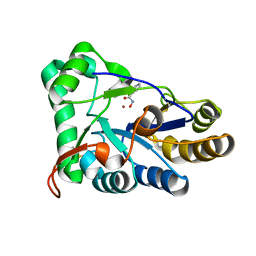 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | BENZHYDROXAMIC ACID, Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
8HFA
 
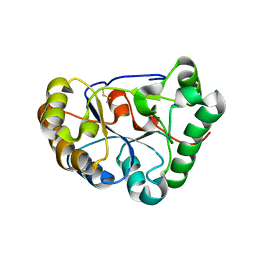 | |
7VQD
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and geranylgeranyl pyrophosphate. | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, NerylNeryl pyrophosphate, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
8HF9
 
 | | The structure of chitin deacetylase Pst_13661 from Puccinia striiformis f. sp. tritici | | Descriptor: | Chitin deacetylase, ZINC ION | | Authors: | Liu, L, Li, Y.C, Zhou, Y, Yang, Q. | | Deposit date: | 2022-11-10 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Inhibition of chitin deacetylases to attenuate plant fungal diseases.
Nat Commun, 14, 2023
|
|
7VQC
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with pyrophosphate | | Descriptor: | Di-trans-poly-cis-decaprenylcistransferase, PYROPHOSPHATE 2-, SULFATE ION | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQ9
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl thiopyrophosphate and isopentyl S-thiolodiphosphate | | Descriptor: | 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQB
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with farnesyl pyrophosphate and dimethylallyl diphosphate | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, FARNESYL DIPHOSPHATE, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7VQA
 
 | | Structure of MA1831 from Methanosarcina acetivorans in complex with dimethylallyl diphosphate. | | Descriptor: | DIMETHYLALLYL DIPHOSPHATE, Di-trans-poly-cis-decaprenylcistransferase, MAGNESIUM ION, ... | | Authors: | Zhang, L.L, Chen, C.C, Liu, W.D, Huang, J.W, Zhang, X.W, Liu, B.B, Guo, R.T. | | Deposit date: | 2021-10-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights to a bi-functional isoprenyl diphosphate synthase that can catalyze head-to-tail and head-to-middle condensation.
Int.J.Biol.Macromol., 214, 2022
|
|
7RVO
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVM
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI11 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVW
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | Descriptor: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RSM
 
 | |
7RVS
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVR
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI18 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI27 | | Descriptor: | 3C-like proteinase, N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
 | |
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVV
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
