7FH0
 
 | |
7QXX
 
 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QY7
 
 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXP
 
 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXU
 
 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXW
 
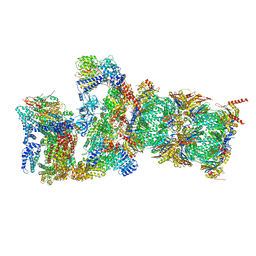 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
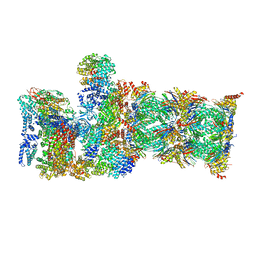 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXN
 
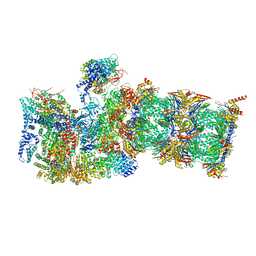 | | Proteasome-ZFAND5 Complex Z+A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
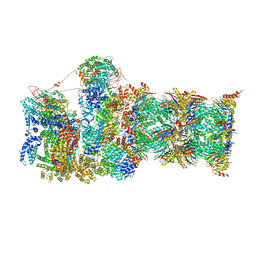 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
4YZ9
 
 | |
4YZC
 
 | | Crystal structure of pIRE1alpha in complex with staurosporine | | Descriptor: | STAUROSPORINE, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
6WUS
 
 | |
4YZD
 
 | | Crystal Structure of human phosphorylated IRE1alpha in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Concha, N.O. | | Deposit date: | 2015-03-24 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Long-Range Inhibitor-Induced Conformational Regulation of Human IRE1 alpha Endoribonuclease Activity.
Mol.Pharmacol., 88, 2015
|
|
7VOA
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with aRBD5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpaca nanobody | | Authors: | Ma, H, Zeng, W.H, Jin, T.C. | | Deposit date: | 2021-10-13 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hetero-bivalent nanobodies provide broad-spectrum protection against SARS-CoV-2 variants of concern including Omicron.
Cell Res., 32, 2022
|
|
5DW3
 
 | |
5DVZ
 
 | | Holo TrpB from Pyrococcus furiosus | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Directed evolution of the tryptophan synthase beta-subunit for stand-alone function recapitulates allosteric activation.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5E0K
 
 | |
6E1C
 
 | |
4D3V
 
 | |
4D3J
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3M
 
 | |
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3N
 
 | |
4D3I
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-((5-(aminomethyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(aminomethyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), GLYCEROL, N-PROPANOL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3T
 
 | |
