1GJ8
 
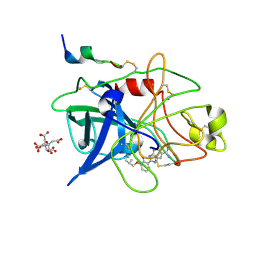 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 6-FLUORO-2-(2-HYDROXY-3-ISOBUTOXY-PHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
6VAR
 
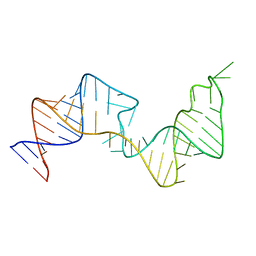 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | Descriptor: | RNA (61-MER) | | Authors: | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
6VJF
 
 | | The P-Loop K to A mutation of C. therm Vps1 GTPase-BSE | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Putative sorting protein Vps1 | | Authors: | Tornabene, B.A, Varlakhanova, N.V, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2020-01-15 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structural and functional characterization of the dominant negative P-loop lysine mutation in the dynamin superfamily protein Vps1.
Protein Sci., 29, 2020
|
|
1HEL
 
 | |
1HEM
 
 | |
1HER
 
 | |
7TIM
 
 | | STRUCTURE OF THE TRIOSEPHOSPHATE ISOMERASE-PHOSPHOGLYCOLOHYDROXAMATE COMPLEX: AN ANALOGUE OF THE INTERMEDIATE ON THE REACTION PATHWAY | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Davenport, R.C, Bash, P.A, Seaton, B.A, Karplus, M, Petsko, G.A, Ringe, D. | | Deposit date: | 1991-04-23 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway.
Biochemistry, 30, 1991
|
|
1HEO
 
 | |
1HEP
 
 | |
1HEN
 
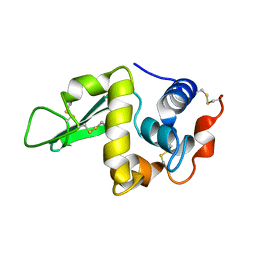 | |
1HEQ
 
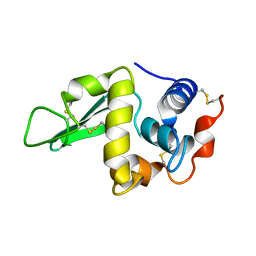 | |
6WMT
 
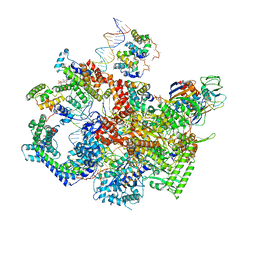 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6VVO
 
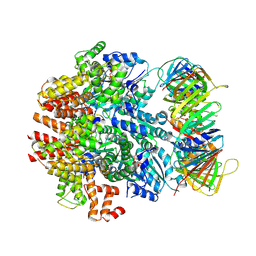 | | Structure of the human clamp loader (Replication Factor C, RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen, PCNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gaubitz, C, Liu, X, Stone, N.P, Kelch, B.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-02-26 | | Last modified: | 2020-03-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the human clamp loader bound to the sliding clamp: a further twist on AAA+ mechanism
Biorxiv, 2020
|
|
6WII
 
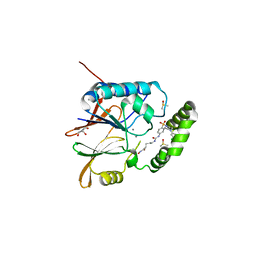 | | Crystal structure of the K. pneumoniae LpxH/JH-LPH-41 complex | | Descriptor: | 5-({[4-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)phenyl]carbamoyl}amino)-N-hydroxypentanamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Cochrane, C.S, Cho, J, Fenton, B.A, Zhou, P. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and evaluation of sulfonyl piperazine LpxH inhibitors.
Bioorg.Chem., 102, 2020
|
|
6VRA
 
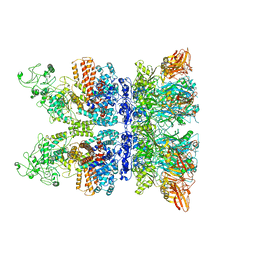 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
6WMP
 
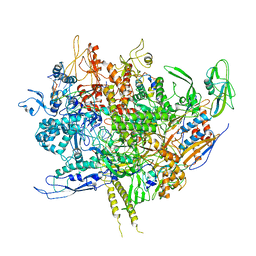 | | F. tularensis RNAPs70-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA T-strand, DNA-directed RNA polymerase subunit alpha 1, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6WMU
 
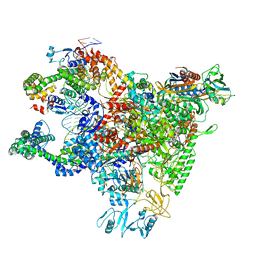 | | E. coli RNAPs70-SspA-gadA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6WJJ
 
 | | Anthrax octamer prechannel bound to full-length lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen, ... | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
6WMR
 
 | | F. tularensis RNAPs70-(MglA-SspA)-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6WY6
 
 | | Crystal structure of S. cerevisiae Atg8 in complex with Ede1 (1220-1247) | | Descriptor: | Autophagy-related protein 8, EH domain-containing and endocytosis protein 1 | | Authors: | Zheng, Y, Wilfling, F, Baumeister, W, Schulman, B.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits.
Mol.Cell, 80, 2020
|
|
5TL4
 
 | | Crystal structure of Sphingomonas paucimobilis aryl O-demethylase LigM | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kohler, A.C, Adams, P.D, Simmons, B.A, Sale, K.L. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Structure of aryl O-demethylase offers molecular insight into a catalytic tyrosine-dependent mechanism.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UDH
 
 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5V88
 
 | | Structure of DCN1 bound to NAcM-COV | | Descriptor: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5VAL
 
 | | BRAF in Complex with N-(3-(tert-butyl)phenyl)-4-methyl-3-(6-morpholinopyrimidin-4-yl)benzamide | | Descriptor: | N-(3-tert-butylphenyl)-4-methyl-3-[6-(morpholin-4-yl)pyrimidin-4-yl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2017-03-27 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design and Discovery of N-(2-Methyl-5'-morpholino-6'-((tetrahydro-2H-pyran-4-yl)oxy)-[3,3'-bipyridin]-5-yl)-3-(trifluoromethyl)benzamide (RAF709): A Potent, Selective, and Efficacious RAF Inhibitor Targeting RAS Mutant Cancers.
J. Med. Chem., 60, 2017
|
|
5VSO
 
 | | NMR structure of Ydj1 J-domain, a cytosolic Hsp40 from Saccharomyces cerevisiae | | Descriptor: | Yeast dnaJ protein 1 | | Authors: | Ciesielski, S.J, Tonelli, M, Lee, W, Cornilescu, G, Markley, J.L, Schilke, B.A, Ziegelhoffer, T, Craig, E.A. | | Deposit date: | 2017-05-12 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Broadening the functionality of a J-protein/Hsp70 molecular chaperone system.
PLoS Genet., 13, 2017
|
|
