8VT5
 
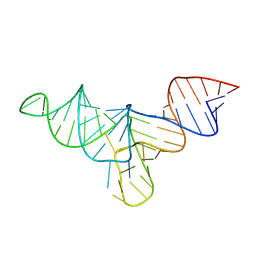 | | Crystal structure of human menRNA | | Descriptor: | menRNA | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2024-01-25 | | Release date: | 2024-07-24 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural basis of NEAT1 lncRNA maturation and menRNA instability.
Nat.Struct.Mol.Biol., 2024
|
|
8VTZ
 
 | |
8VU0
 
 | |
8V1I
 
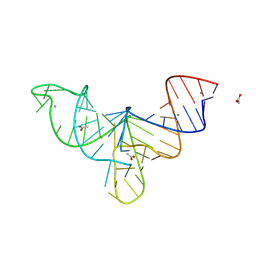 | | Crystal structure of human mascRNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Skeparnias, I, Zhang, J. | | Deposit date: | 2023-11-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of MALAT1 RNA maturation and mascRNA biogenesis.
Nat.Struct.Mol.Biol., 2024
|
|
8GJM
 
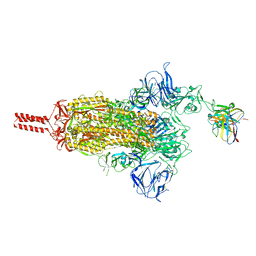 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
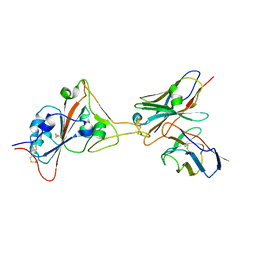 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8HVU
 
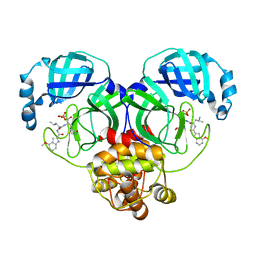 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVV
 
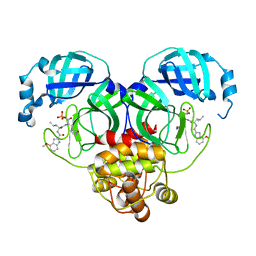 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVX
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVZ
 
 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVW
 
 | | Crystal structure of SARS-Cov-2 main protease M49I mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zou, X.F, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG5
 
 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG7
 
 | |
8IG9
 
 | |
8IG8
 
 | |
8IGB
 
 | |
5GO0
 
 | |
5HNK
 
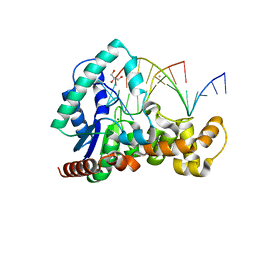 | | Crystal structure of T5Fen in complex intact substrate and metal ions. | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*GP*CP*GP*TP*AP*CP*GP*C)-3'), Exodeoxyribonuclease, GLYCEROL, ... | | Authors: | Almalki, F.A, Feng, M, Zhang, J, Sedelnikova, S.E, Rafferty, J.B, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Direct observation of DNA threading in flap endonuclease complexes.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7XJF
 
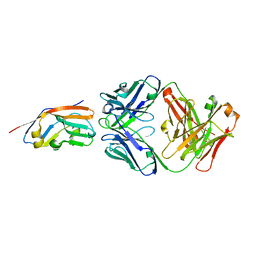 | | Crystal structure of 6MW3211 Fab in complex with CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wang, J, Wang, R, Jiao, S, Wang, S, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2022-04-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Theranostics, 13, 2023
|
|
5HRB
 
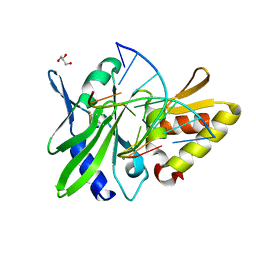 | | The crystal structure of AsfvPolX:DNA1 binary complex | | Descriptor: | BETA-MERCAPTOETHANOL, DNA (5'-D(*CP*GP*GP*AP*TP*AP*TP*CP*C)-3'), DNA polymerase beta-like protein, ... | | Authors: | Chen, Y.Q, Zhang, J, Gan, J.H. | | Deposit date: | 2016-01-23 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of Se-AsfvPolX(L52/163M mutant) in complex with 1nt-gap DNA1
To Be Published
|
|
5HRH
 
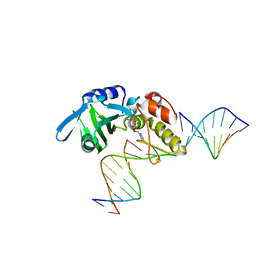 | |
5HRD
 
 | |
5HRK
 
 | |
5HRF
 
 | |
