1R5I
 
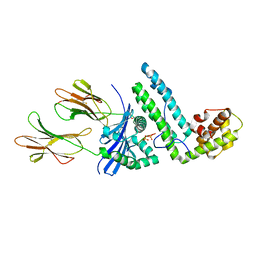 | | Crystal structure of the MAM-MHC complex | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Zhao, Y, Li, Z, Drozd, S.J, Guo, Y, Mourad, W, Li, H. | | Deposit date: | 2003-10-10 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Mycoplasma arthritidis mitogen complexed with HLA-DR1 reveals a novel superantigen fold and a dimerized superantigen-MHC complex.
Structure, 12, 2004
|
|
3JUG
 
 | |
4ZFC
 
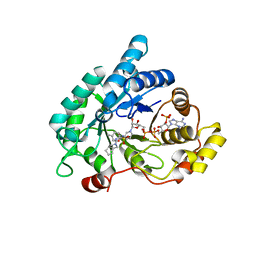 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVV
 
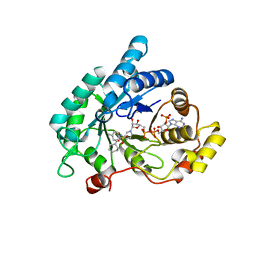 | | Crystal structure of AKR1C3 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
 | | Crystal structure of AKR1C3 complexed with glimepiride | | Descriptor: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVP
 
 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
1ONS
 
 | |
4CNC
 
 | | Crystal structure of human 5T4 (Wnt-activated inhibitory factor 1, Trophoblast glycoprotein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhao, Y, Malinauskas, T, Harlos, K, Jones, E.Y. | | Deposit date: | 2014-01-21 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Insights Into the Inhibition of Wnt Signaling by Cancer Antigen 5T4/Wnt-Activated Inhibitory Factor 1.
Structure, 22, 2014
|
|
4CNM
 
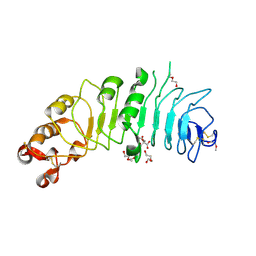 | | Crystal structure of human 5T4 (Wnt-activated inhibitory factor 1, Trophoblast glycoprotein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Zhao, Y, Malinauskas, T, Harlos, K, Jones, E.Y. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Inhibition of Wnt Signaling by Cancer Antigen 5T4/Wnt-Activated Inhibitory Factor 1.
Structure, 22, 2014
|
|
4CYH
 
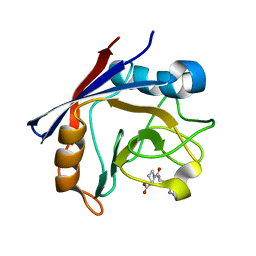 | | CYCLOPHILIN A COMPLEXED WITH DIPEPTIDE HIS-PRO | | Descriptor: | CYCLOPHILIN A, HISTIDINE, PROLINE | | Authors: | Zhao, Y, Ke, H. | | Deposit date: | 1996-02-27 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic implication of crystal structures of the cyclophilin-dipeptide complexes.
Biochemistry, 35, 1996
|
|
5K4F
 
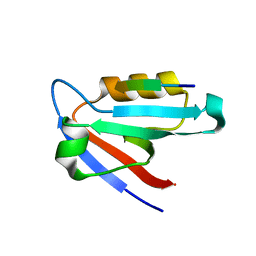 | | CAL PDZ mutant C319A with a peptide | | Descriptor: | Golgi-associated PDZ and coiled-coil motif-containing protein, HPV18E6 peptide | | Authors: | Zhao, Y, Madden, D.R. | | Deposit date: | 2016-05-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CAL PDZ domain mutant with a peptide
To Be Published
|
|
5JQ7
 
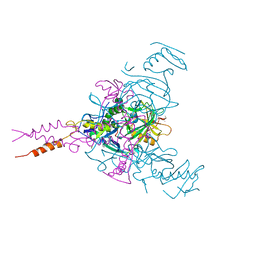 | | Crystal structure of Ebola glycoprotein in complex with toremifene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
6TV4
 
 | | CFF-Notum complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CAFFEINE, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-01-08 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Caffeine inhibits Notum activity by binding at the catalytic pocket.
Commun Biol, 3, 2020
|
|
5KK2
 
 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
6ZYF
 
 | | Notum_Ghrelin complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2020-07-31 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Notum deacylates octanoylated ghrelin.
Mol Metab, 49, 2021
|
|
4DL2
 
 | | Human DNA polymerase eta inserting dCMPNPP opposite CG template (GG0a) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*CP*GP*GP*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DL7
 
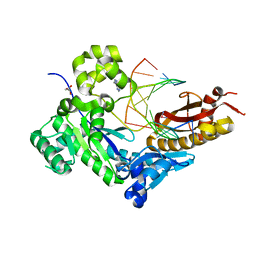 | | Human DNA polymerase eta fails to extend primer 2 nucleotide after cisplatin crosslink (Pt-GG4). | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Cisplatin, DNA (5'-D(*TP*AP*CP*TP*CP*GP*GP*TP*CP*AP*CP*T)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7R9M
 
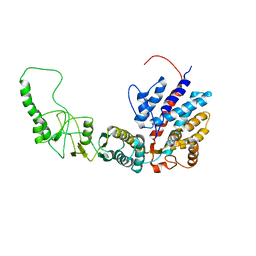 | | Methanococcus maripaludis chaperonin, closed conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9E
 
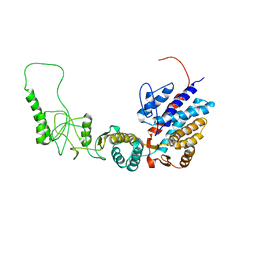 | | Methanococcus maripaludis chaperonin, open conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7RAK
 
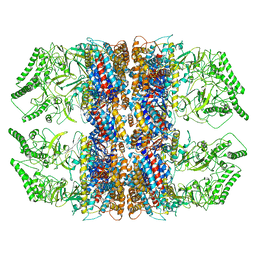 | | Methanococcus maripaludis chaperonin complex in open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9O
 
 | | Methanococcus maripaludis chaperonin, closed conformation 1 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9U
 
 | | Methanococcus maripaludis chaperonin, closed conformation 3 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9J
 
 | | Methanococcus maripaludis chaperonin, open conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9I
 
 | | Methanococcus maripaludis chaperonin, open conformation 2 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
7R9K
 
 | | Methanococcus maripaludis chaperonin, closed conformation 4 | | Descriptor: | Chaperonin | | Authors: | Zhao, Y, Schmid, M, Frydman, J, Chiu, W. | | Deposit date: | 2021-06-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | CryoEM reveals the stochastic nature of individual ATP binding events in a group II chaperonin.
Nat Commun, 12, 2021
|
|
