6IXJ
 
 | | The crystal structure of sulfoacetaldehyde reductase from Klebsiella oxytoca | | Descriptor: | 2-hydroxyethylsulfonic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sulfoacetaldehyde reductase | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural investigation of sulfoacetaldehyde reductase fromKlebsiella oxytoca.
Biochem. J., 476, 2019
|
|
6J6P
 
 | | Crystal structure of diamondback moth ryanodine receptor phosphorylation domain(2836-3050) mutant S2946D | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Xu, T, Lin, L, Yuchi, Z. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of diamondback moth ryanodine receptor Repeat34 domain reveals insect-specific phosphorylation sites.
Bmc Biol., 17, 2019
|
|
6JKP
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense in complex with NAD+ | | Descriptor: | Methanol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.008 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
6JKO
 
 | | Crystal structure of sulfoacetaldehyde reductase from Bifidobacterium kashiwanohense | | Descriptor: | Methanol dehydrogenase, ZINC ION | | Authors: | Zhou, Y, Xu, T, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and characterization of a new sulfoacetaldehyde reductase from the human gut bacteriumBifidobacterium kashiwanohense.
Biosci.Rep., 39, 2019
|
|
7E12
 
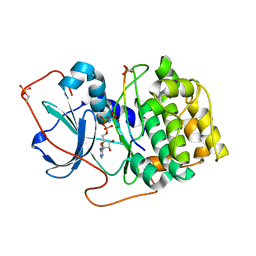 | | Crystal structure of PKAc-A11E complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, THR-ARG-SER-GLU-ILE-ARG-ARG-ALA-SER-THR-ILE-GLU, ... | | Authors: | Qin, J, Lin, L, Yuchi, Z. | | Deposit date: | 2021-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structures of PKA-phospholamban complexes reveal a mechanism of familial dilated cardiomyopathy.
Elife, 11, 2022
|
|
6JIX
 
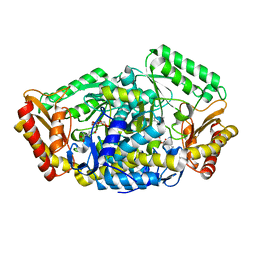 | | The cyrstal structure of taurine:2-oxoglutarate aminotransferase from Bifidobacterium kashiwanohense, in complex with PLP and glutamate | | Descriptor: | GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE, taurine:2-oxoglutarate aminotransferase | | Authors: | Li, M, Lin, L, Zhang, Y, Yuchi, Z. | | Deposit date: | 2019-02-23 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Biochemical and structural investigation of taurine:2-oxoglutarate aminotransferase fromBifidobacterium kashiwanohense.
Biochem.J., 476, 2019
|
|
6KIM
 
 | |
6J6O
 
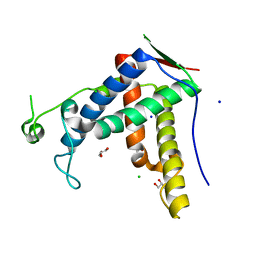 | | Crystal structure of diamondback moth ryanodine receptor phosphorylation domain(2836-3050) | | Descriptor: | CHLORIDE ION, GLYCEROL, Ryanodine receptor, ... | | Authors: | Xu, T, Lin, L, Yuchi, Z. | | Deposit date: | 2019-01-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Crystal structure of diamondback moth ryanodine receptor Repeat34 domain reveals insect-specific phosphorylation sites.
Bmc Biol., 17, 2019
|
|
7VUA
 
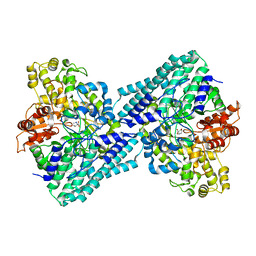 | | Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme | | Descriptor: | (4S)-4-hydroxy-D-proline, HplG | | Authors: | Duan, Y, Lu, Q, Yuchi, Z, Zhang, Y. | | Deposit date: | 2021-11-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Anaerobic Hydroxyproline Degradation Involving C-N Cleavage by a Glycyl Radical Enzyme.
J.Am.Chem.Soc., 144, 2022
|
|
7E7L
 
 | | The crystal structure of arylacetate decarboxylase from Olsenella scatoligenes. | | Descriptor: | 4-HYDROXYPHENYLACETATE, Hydroxyphenylacetic acid decarboxylase | | Authors: | Lu, Q, Duan, Y, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | The Glycyl Radical Enzyme Arylacetate Decarboxylase from Olsenella scatoligenes
Acs Catalysis, 11, 2021
|
|
7EEV
 
 | | Structure of UTP cyclohydrolase | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, GTP cyclohydrolase II, ZINC ION | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-07-28 | | Last modified: | 2022-09-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
7EJ3
 
 | | UTP cyclohydrolase | | Descriptor: | DIPHOSPHATE, GLYCINE, GTP cyclohydrolase II, ... | | Authors: | Zhang, H, Zhang, Y, Yuchi, Z. | | Deposit date: | 2021-04-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical investigation of UTP cyclohydrolase
Acs Catalysis, 2021
|
|
7CF9
 
 | | Structure of RyR1 (Ca2+/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, CALCIUM ION, Peptidyl-prolyl cis-trans isomerase FKBP1B, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-06-24 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
6MM5
 
 | |
6MM7
 
 | |
6MM6
 
 | |
6MM8
 
 | |
5ZXL
 
 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
7F2B
 
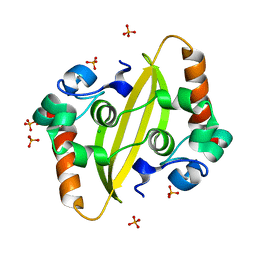 | |
7F2E
 
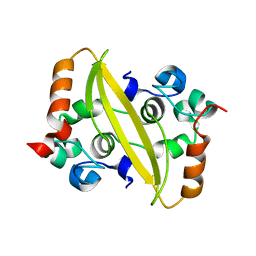 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
7WJM
 
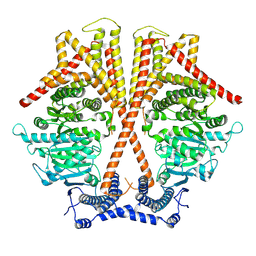 | |
7WJO
 
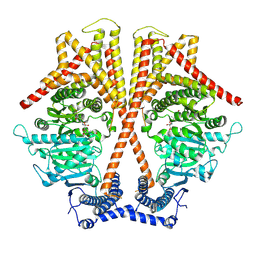 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with nikkomycin Z | | Descriptor: | (2S)-{[(2S,3S,4S)-2-amino-4-hydroxy-4-(5-hydroxypyridin-2-yl)-3-methylbutanoyl]amino}[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]acetic acid (non-preferred name), Chitin synthase | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7WJN
 
 | | CryoEM structure of chitin synthase 1 mutant E495A from Phytophthora sojae complexed with UDP-GlcNAc | | Descriptor: | Chitin synthase, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7X05
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with the nascent chitooligosaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin synthase, MANGANESE (II) ION, ... | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
7X06
 
 | | CryoEM structure of chitin synthase 1 from Phytophthora sojae complexed with UDP | | Descriptor: | Chitin synthase, MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Chen, W, Cao, P, Gong, Y, Yang, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for directional chitin biosynthesis.
Nature, 610, 2022
|
|
