8QYV
 
 | | SWR1-hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
1CKN
 
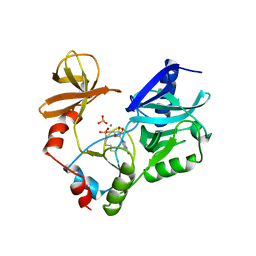 | | STRUCTURE OF GUANYLYLATED MRNA CAPPING ENZYME COMPLEXED WITH GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, MRNA CAPPING ENZYME, ... | | Authors: | Hakansson, K, Doherty, A.J, Wigley, D.B. | | Deposit date: | 1997-04-20 | | Release date: | 1997-07-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes.
Cell(Cambridge,Mass.), 89, 1997
|
|
1CKM
 
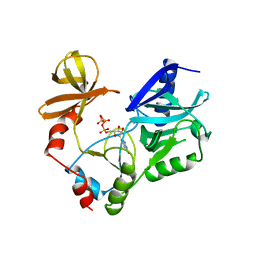 | |
1W5T
 
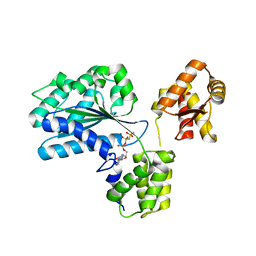 | | Structure of the Aeropyrum Pernix ORC2 protein (ADPNP-ADP complexes) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ORC2, ... | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
1W5S
 
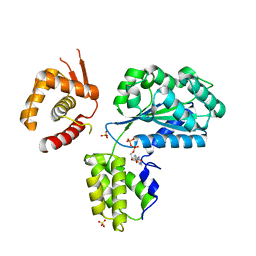 | | Structure of the Aeropyrum Pernix ORC2 protein (ADP form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ORIGIN RECOGNITION COMPLEX SUBUNIT 2 ORC2, SULFATE ION | | Authors: | Singleton, M.R, Morales, R, Grainge, I, Cook, N, Isupov, M.N, Wigley, D.B. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes Induced by Nucleotide Binding in Cdc6/Orc from Aeropyrum Pernix
J.Mol.Biol., 343, 2004
|
|
1W36
 
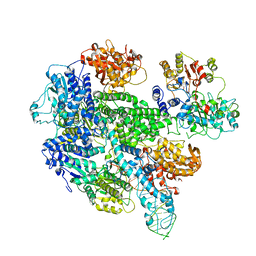 | | RecBCD:DNA complex | | Descriptor: | CALCIUM ION, DNA HAIRPIN, EXODEOXYRIBONUCLEASE V ALPHA CHAIN, ... | | Authors: | Singleton, M.R, Dillingham, M.S, Gaudier, M, C Kowalczykowski, S, Wigley, D.B. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Recbcd Enzyme Reveals a Machine for Processing DNA Breaks
Nature, 432, 2004
|
|
1OTF
 
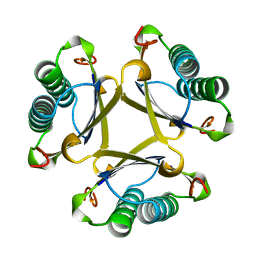 | | 4-OXALOCROTONATE TAUTOMERASE-TRICLINIC CRYSTAL FORM | | Descriptor: | 4-OXALOCROTONATE TAUTOMERASE | | Authors: | Subramanya, H.S, Roper, D.I, Dauter, Z, Dodson, E.J, Davies, G.J, Wilson, K.S, Wigley, D.B. | | Deposit date: | 1995-11-09 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases.
Biochemistry, 35, 1996
|
|
6FWR
 
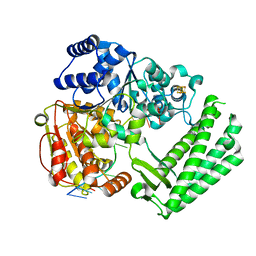 | | Structure of DinG in complex with ssDNA | | Descriptor: | ATP-dependent DNA helicase DinG, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), IRON/SULFUR CLUSTER | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2018-03-07 | | Release date: | 2018-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA translocation mechanism of an XPD family helicase.
Elife, 7, 2018
|
|
6GEN
 
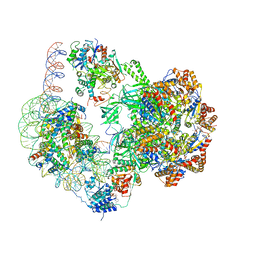 | | Chromatin remodeller-nucleosome complex at 4.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
6GEJ
 
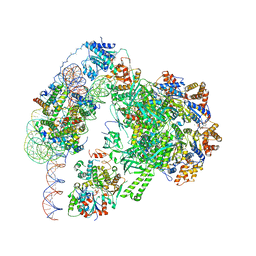 | | Chromatin remodeller-nucleosome complex at 3.6 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-26 | | Release date: | 2018-10-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
1IGD
 
 | |
4AM6
 
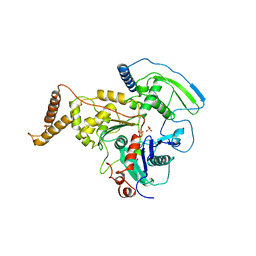 | | C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, SULFATE ION | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AM7
 
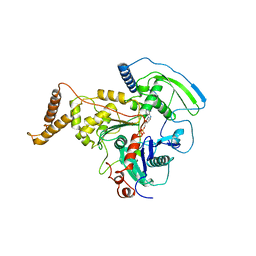 | | ADP-BOUND C-TERMINAL DOMAIN OF ACTIN-RELATED PROTEIN ARP8 FROM S. CEREVISIAE | | Descriptor: | ACTIN-LIKE PROTEIN ARP8, ADENOSINE-5'-DIPHOSPHATE | | Authors: | Wuerges, J, Saravanan, M, Bose, D, Cook, N.J, Zhang, X, Wigley, D.B. | | Deposit date: | 2012-03-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interactions between the Nucleosome Histone Core and Arp8 in the Ino80 Chromatin Remodeling Complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2CHV
 
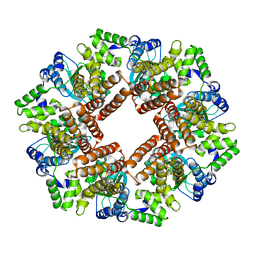 | | Replication Factor C ADPNP complex | | Descriptor: | REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CHQ
 
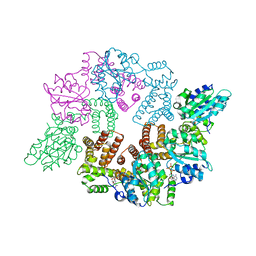 | | Replication Factor C ADPNP complex | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
2CHG
 
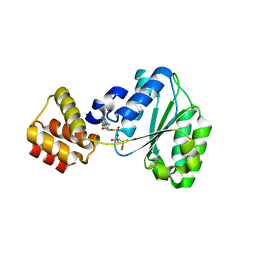 | | Replication Factor C domains 1 and 2 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
6QXT
 
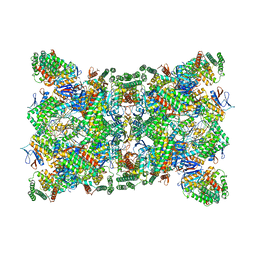 | | Cas1-Cas2-Csn2-DNA dimer complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
6QY3
 
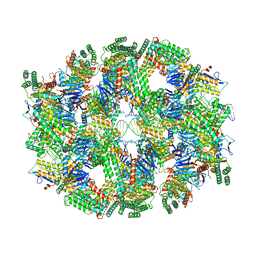 | | Segment of the Cas1-Cas2-Csn2-DNA filament complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-08 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
6SER
 
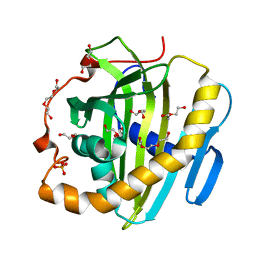 | | Crystal structure of human STARD10 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, START domain-containing protein 10, ... | | Authors: | Cheng, K, Wigley, D.B. | | Deposit date: | 2019-07-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | The crystal structure of human STARD10
To Be Published
|
|
6SJF
 
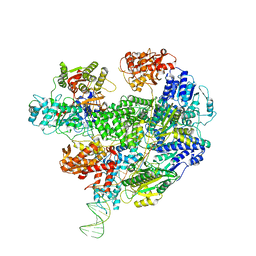 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJG
 
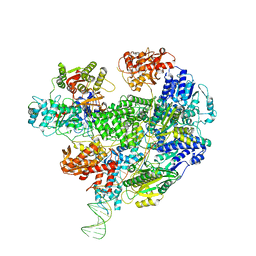 | | Cryo-EM structure of the RecBCD no Chi negative control complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6QXF
 
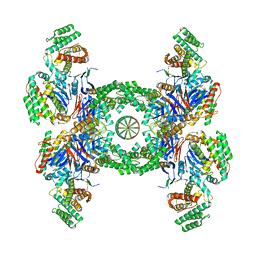 | | Cas1-Cas2-Csn2-DNA complex from the Type II-A CRISPR-Cas system | | Descriptor: | CALCIUM ION, CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, ... | | Authors: | Wilkinson, M, Drabavicius, G, Silanskas, A, Gasiunas, G, Siksnys, V, Wigley, D.B. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of the DNA-Bound Spacer Capture Complex of a Type II CRISPR-Cas System.
Mol.Cell, 75, 2019
|
|
6SJE
 
 | | Cryo-EM structure of the RecBCD Chi partially-recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SJB
 
 | | Cryo-EM structure of the RecBCD Chi recognised complex | | Descriptor: | DNA fork substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T2U
 
 | | Cryo-EM structure of the RecBCD in complex with Chi-minus2 substrate | | Descriptor: | DNA (Chi-minus2), RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-10-09 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
