1RLO
 
 | | Phospho-aspartyl Intermediate Analogue of ybiV from E. coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1PUX
 
 | | NMR Solution Structure of BeF3-Activated Spo0F, 20 conformers | | Descriptor: | Sporulation initiation phosphotransferase F | | Authors: | Gardino, A.K, Volkman, B.F, Cho, H.S, Lee, S.Y, Wemmer, D.E, Kern, D. | | Deposit date: | 2003-06-25 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of BeF(3)(-)-activated Spo0F reveals the conformational switch in a phosphorelay system.
J.Mol.Biol., 331, 2003
|
|
1RLT
 
 | | Transition State Analogue of ybiV from E. coli K12 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1RLM
 
 | | Crystal Structure of ybiV from Escherichia coli K12 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphatase | | Authors: | Roberts, A, Lee, S.Y, McCullagh, E, Silversmith, R.E, Wemmer, D.E. | | Deposit date: | 2003-11-26 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ybiv from Escherichia coli K12 is a HAD phosphatase.
Proteins, 58, 2005
|
|
1PQQ
 
 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1TM9
 
 | | NMR Structure of gene target number gi3844938 from Mycoplasma genitalium: Berkeley Structural Genomics Center | | Descriptor: | Hypothetical protein MG354 | | Authors: | Pelton, J.G, Shi, J, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Gene Target gi3844938 from Mycoplasma genitalium
To be Published
|
|
1SXL
 
 | |
1DB6
 
 | | SOLUTION STRUCTURE OF THE DNA APTAMER 5'-CGACCAACGTGTCGCCTGGTCG-3' COMPLEXED WITH ARGININAMIDE | | Descriptor: | ARGININEAMIDE, DNA | | Authors: | Robertson, S.A, Harada, K, Frankel, A.D, Wemmer, D.E. | | Deposit date: | 1999-11-02 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination and binding kinetics of a DNA aptamer-argininamide complex.
Biochemistry, 39, 2000
|
|
1FO5
 
 | | SOLUTION STRUCTURE OF REDUCED MJ0307 | | Descriptor: | THIOREDOXIN | | Authors: | Cave, J.W, Cho, H.S, Batchelder, A.M, Kim, R, Yokota, H, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-08-24 | | Release date: | 2001-04-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii.
Protein Sci., 10, 2001
|
|
1ZDM
 
 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
1BBI
 
 | |
2KIC
 
 | | n-NafY. N-terminal domain of NafY | | Descriptor: | Nitrogenase gamma subunit | | Authors: | Phillips, A.H, Hernandez, J.A, Erbil, K, Pelton, J.G, Wemmer, D.E, Rubio, L.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biological activity and solution structure of the apo-dinitrogenase binding domain of NafY
J.Biol.Chem., 2010
|
|
2LAI
 
 | |
2JMP
 
 | |
2MXD
 
 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
2O8K
 
 | | NMR Structure of the Sigma-54 RpoN Domain Bound to the-24 Promoter Element | | Descriptor: | 5'-D(*GP*AP*AP*AP*CP*GP*TP*GP*CP*CP*AP*AP*AP*A)-3', 5'-D(*TP*TP*TP*TP*GP*GP*CP*AP*CP*GP*TP*TP*TP*C)-3', RNA polymerase sigma factor RpoN | | Authors: | Doucleff, M, Pelton, J.G, Lee, P.S, Wemmer, D.E. | | Deposit date: | 2006-12-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA recognition by the alternative sigma-factor, sigma54.
J.Mol.Biol., 369, 2007
|
|
108D
 
 | | THE SOLUTION STRUCTURE OF A DNA COMPLEX WITH THE FLUORESCENT BIS INTERCALATOR TOTO DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | 1,1-(4,4,8,8-TETRAMETHYL-4,8-DIAZAUNDECAMETHYLENE)-BIS-4-3-METHYL-2,3-DIHYDRO-(BENZO-1,3-THIAZOLE)-2-METHYLIDENE)-QUINOLINIUM, DNA (5'-D(*CP*GP*CP*TP*AP*GP*CP*G)-3') | | Authors: | Spielmann, H.P, Wemmer, D.E, Jacobsen, J.P. | | Deposit date: | 1995-01-31 | | Release date: | 1995-06-03 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA complex with the fluorescent bis-intercalator TOTO determined by NMR spectroscopy.
Biochemistry, 34, 1995
|
|
1J56
 
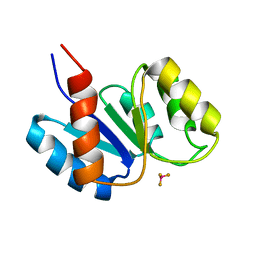 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRX
 
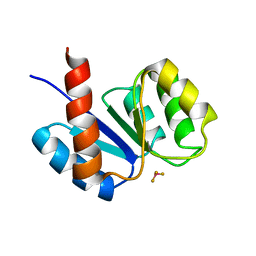 | | SOLUTION STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURES INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
1KRW
 
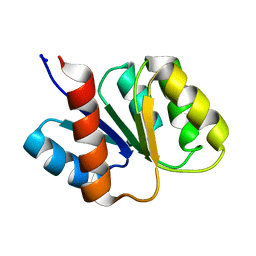 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
204D
 
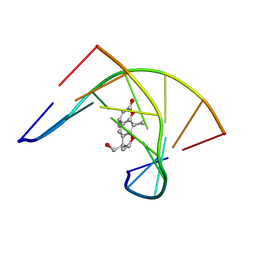 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
203D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
2BBI
 
 | |
1LLS
 
 | | CRYSTAL STRUCTURE OF UNLIGANDED MALTOSE BINDING PROTEIN WITH XENON | | Descriptor: | Maltose-binding periplasmic protein, XENON | | Authors: | Rubin, S.M, Lee, S.-Y, Ruiz, E.J, Pines, A, Wemmer, D.E. | | Deposit date: | 2002-04-30 | | Release date: | 2002-09-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DETECTION AND CHARACTERIZATION OF XENON-BINDING SITES IN PROTEINS BY 129XE NMR SPECTROSCOPY
J.MOL.BIOL., 322, 2002
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | Descriptor: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | Authors: | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | Deposit date: | 2001-03-19 | | Release date: | 2001-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
