2ZD0
 
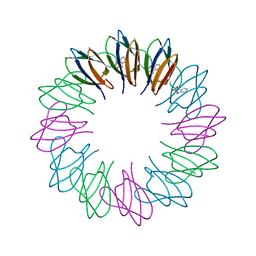 | | Crystal structures and thermostability of mutant TRAP3 A5 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2E89
 
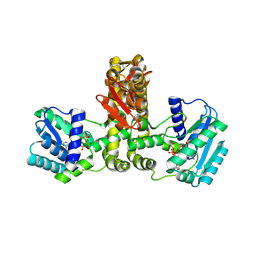 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
2ZP8
 
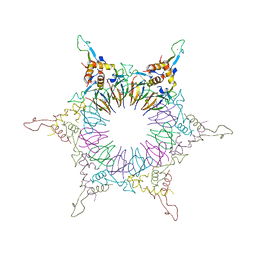 | | The Nature of the TRAP:Anti-TRAP complex | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB, Tryptophan RNA-binding attenuator protein-inhibitory protein, ... | | Authors: | Watanabe, M, Heddle, J.G, Unzai, S, Akashi, S, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The nature of the TRAP-Anti-TRAP complex.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2CYA
 
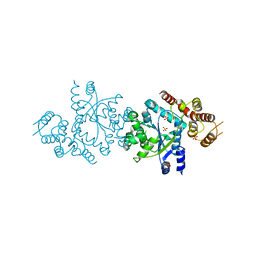 | | Crystal structure of tyrosyl-tRNA synthetase from Aeropyrum pernix | | Descriptor: | SULFATE ION, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Kuroishi, C, Kuramitsu, S, Terada, T, Bessho, Y, Shirouzu, M, Sekine, S.I, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2005
|
|
2CYC
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Pyrococcus horikoshii | | Descriptor: | TYROSINE, tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Sakamoto, K, Terada, T, Shirouzu, M, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
2LTQ
 
 | | High resolution structure of DsbB C41S by joint calculation with solid-state NMR and X-ray data | | Descriptor: | Disulfide bond formation protein B, Fab fragment heavy chain, Fab fragment light chain, ... | | Authors: | Tang, M, Sperling, L.J, Schwieters, C.D, Nesbitt, A.E, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Structure of the Disulfide Bond Generating Membrane Protein DsbB in the Lipid Bilayer.
J.Mol.Biol., 425, 2013
|
|
2CYB
 
 | | Crystal structure of Tyrosyl-tRNA Synthetase complexed with L-tyrosine from Archaeoglobus fulgidus | | Descriptor: | TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kuratani, M, Sakai, H, Takahashi, M, Yanagisawa, T, Kobayashi, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-06 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Tyrosyl-tRNA Synthetases from Archaea
J.Mol.Biol., 355, 2006
|
|
1WWR
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Aquifex aeolicus | | Descriptor: | ZINC ION, tRNA adenosine deaminase TadA | | Authors: | Kuratani, M, Ishii, R, Bessho, Y, Fukunaga, R, Sengoku, T, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-12 | | Release date: | 2005-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of tRNA Adenosine Deaminase (TadA) from Aquifex aeolicus
J.Biol.Chem., 280, 2005
|
|
2ZCZ
 
 | | Crystal structures and thermostability of mutant TRAP3 A7 (ENGINEERED TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Watanabe, M, Mishima, Y, Yamashita, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Intersubunit linker length as a modifier of protein stability: crystal structures and thermostability of mutant TRAP.
Protein Sci., 17, 2008
|
|
2LEG
 
 | | Membrane protein complex DsbB-DsbA structure by joint calculations with solid-state NMR and X-ray experimental data | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein DsbA, UBIQUINONE-1, ... | | Authors: | Tang, M, Sperling, L.J, Berthold, D.A, Schwieters, C.D, Nesbitt, A.E, Nieuwkoop, A.J, Gennis, R.B, Rienstra, C.M. | | Deposit date: | 2011-06-15 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution membrane protein structure by joint calculations with solid-state NMR and X-ray experimental data.
J.Biomol.Nmr, 51, 2011
|
|
4EBW
 
 | | Structure of Focal Adhesion Kinase catalytic domain in complex with novel allosteric inhibitor | | Descriptor: | 1-ethyl-8-(4-ethylphenyl)-5-methyl-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazine 4,4-dioxide, Focal adhesion kinase 1 | | Authors: | Iwatani, M, Iwata, H, Okabe, A, Skene, R.J, Tomita, N, Hayashi, Y, Aramaki, Y, Hosfield, D.J, Hori, A, Baba, A, Miki, H. | | Deposit date: | 2012-03-25 | | Release date: | 2012-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel allosteric FAK inhibitors.
Eur.J.Med.Chem., 61, 2013
|
|
5B5S
 
 | |
2D39
 
 | | Trivalent Recognition Unit of Innate Immunity System; Crystal Structure of human M-ficolin Fibrinogen-like Domain | | Descriptor: | CALCIUM ION, Ficolin-1 | | Authors: | Tanio, M, Kondo, S, Sugio, S, Kohno, T. | | Deposit date: | 2005-09-26 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trivalent recognition unit of innate immunity system; crystal structure of trimeric human m-ficolin fibrinogen-like domain
J.Biol.Chem., 282, 2007
|
|
2YY8
 
 | |
7ED6
 
 | | Crystal structure of Thermus thermophilus FakA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | Authors: | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92850327 Å) | | Cite: | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
7ED9
 
 | | Crystal structure of selenomethionine-labeled Thermus thermophilus FakA ATP-binding domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable kinase | | Authors: | Nakatani, M, Nakahara, S, Fukui, K, Murakawa, T, Masui, R. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01764154 Å) | | Cite: | Crystal structure of a nucleotide-binding domain of fatty acid kinase FakA from Thermus thermophilus HB8.
J.Struct.Biol., 214, 2022
|
|
2Z23
 
 | | Crystal structure of Y.pestis oligo peptide binding protein OppA with tri-lysine ligand | | Descriptor: | Periplasmic oligopeptide-binding protein, peptide (LYS)(LYS)(LYS) | | Authors: | Tanabe, M, Bertland, T, Mirza, O, Byrne, B, Brown, K.A. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3A2S
 
 | |
3AJE
 
 | |
2Z22
 
 | | Crystal structure of phosphate preplasmic binding protein psts from yersinia pestis | | Descriptor: | PHOSPHATE ION, Periplasmic phosphate-binding protein | | Authors: | Tanabe, M, Byrne, B, Brown, K.A, Mirza, O, Bertland, T. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3VZT
 
 | |
3VZW
 
 | |
3VZU
 
 | |
4U74
 
 | | Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (G262A mutant) | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-PHENYL-1H-IMIDAZOLE, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Sugimoto, H, Horitani, M, Kometani, E, Shiro, Y. | | Deposit date: | 2014-07-30 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Conformation and Mobility of Active Site Loop is Critical for Substrate Binding and Inhibition in Human Indoleamine 2,3-Dioxygenase
to be published
|
|
3LQB
 
 | | Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio | | Descriptor: | 1,2-ETHANEDIOL, LOC792177 protein, SULFATE ION, ... | | Authors: | Tanokura, M, Okada, A, Nagata, K, Yasumasu, S, Ohtsuka, J, Iuchi, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of zebrafish hatching enzyme 1 from the zebrafish Danio rerio
J.Mol.Biol., 402, 2010
|
|
