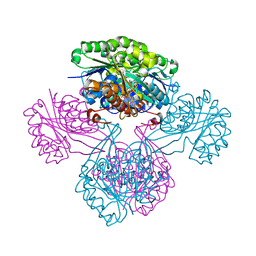
1V1A
 
 | |
1V1S
 
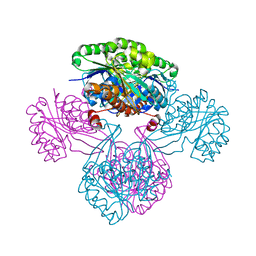 | |
2CVZ
 
 | |
1V7R
 
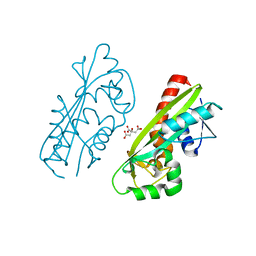 | |
1WR8
 
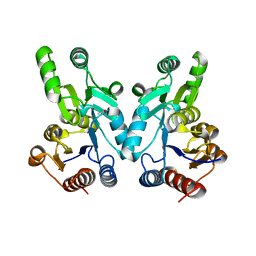 | |
2DLF
 
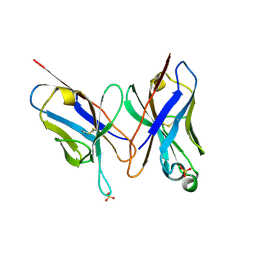 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 6.75 | | Descriptor: | PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S) (HEAVY CHAIN)), PROTEIN (ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S)-KAPPA (LIGHT CHAIN)), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-12-17 | | Release date: | 1999-12-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
1WP4
 
 | |
3GFL
 
 | |
3GF2
 
 | |
3GEZ
 
 | |
3GFI
 
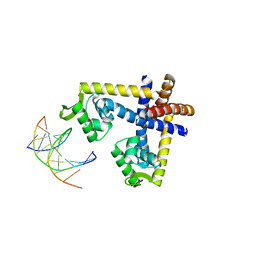 | | Crystal structure of ST1710 complexed with its promoter DNA | | Descriptor: | 146aa long hypothetical transcriptional regulator, 5'-D(*TP*AP*AP*CP*AP*AP*TP*AP*GP*CP*AP*AP*A)-3', 5'-D(*TP*TP*GP*CP*TP*AP*TP*TP*GP*T)-3' | | Authors: | Kumarevel, T, Tanaka, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-02-26 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | ST1710-DNA complex crystal structure reveals the DNA binding mechanism of the MarR family of regulators.
Nucleic Acids Res., 37, 2009
|
|
3GFM
 
 | |
3GFJ
 
 | |
2CYP
 
 | |
