7WOT
 
 | | Cryo-EM structure of the inner ring monomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7WOO
 
 | | Cryo-EM structure of the inner ring protomer of the Saccharomyces cerevisiae nuclear pore complex | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP157, ... | | Authors: | Li, Z.Q, Chen, S.J.B, Zhao, L, Sui, S.F. | | Deposit date: | 2022-01-22 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex.
Cell Res., 32, 2022
|
|
7E8T
 
 | | Monomer of Ypt32-TRAPPII | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
5Y6P
 
 | | Structure of the phycobilisome from the red alga Griffithsia pacifica | | Descriptor: | ApcD, ApcF, LC, ... | | Authors: | Zhang, J, Ma, J.F, Liu, D.S, Sun, S, Sui, S.F. | | Deposit date: | 2017-08-13 | | Release date: | 2017-11-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of phycobilisome from the red alga Griffithsia pacifica
Nature, 551, 2017
|
|
7VC9
 
 | | Tom20 subunits | | Descriptor: | Mitochondrial import receptor subunit TOM20 homolog | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-02 | | Release date: | 2022-09-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Structural basis of Tom20 and Tom22 cytosolic domains as the human TOM complex receptors.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7VBY
 
 | | Tom core complex with Tom20 and Tom22 subunits. | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Tom core complex with Tom20 and Tom22 subunits.
To Be Published
|
|
7VC4
 
 | | Tom complex with Tom22 and Tom20 subunits | | Descriptor: | Mitochondrial import receptor subunit TOM22 homolog, Mitochondrial import receptor subunit TOM40 homolog, Mitochondrial import receptor subunit TOM5 homolog, ... | | Authors: | Liu, D.S, Sui, S.F. | | Deposit date: | 2021-09-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Tom complex with Tom22 and Tom20 subunits
To Be Published
|
|
7VZG
 
 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the larger form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c.
Nat Commun, 13, 2022
|
|
7VZR
 
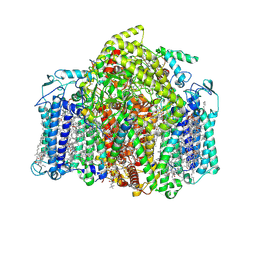 | | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c (the smaller form) | | Descriptor: | BACTERIOCHLOROPHYLL A, CALCIUM ION, CHLOROPHYLL A, ... | | Authors: | Huang, G.Q, Dong, S.S, Qin, X.C, Sui, S.F. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structure of the Acidobacteria homodimeric reaction center bound with cytochrome c
Nat Commun, 13, 2022
|
|
7E94
 
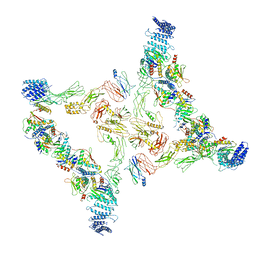 | | Intact TRAPPII (State II) | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7EA3
 
 | | Intact Ypt32-TRAPPII (dimer). | | Descriptor: | GTP-binding protein YPT32/YPT11, TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-06 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E8S
 
 | | Intact TRAPPII (state I). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-02 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
7E93
 
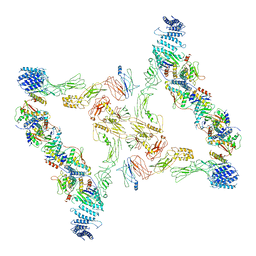 | | Intact TRAPPII (state III). | | Descriptor: | TRAPP-associated protein TCA17, Trafficking protein particle complex II-specific subunit 120, Trafficking protein particle complex II-specific subunit 130, ... | | Authors: | Mi, C.C, Sui, S.F. | | Deposit date: | 2021-03-03 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (6.54 Å) | | Cite: | Structural basis for assembly of TRAPPII complex and specific activation of GTPase Ypt31/32.
Sci Adv, 8, 2022
|
|
6JLU
 
 | | Structure of PSII-FCP supercomplex from a centric diatom Chaetoceros gracilis at 3.02 angstrom resolution | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'-yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pi, X, Zhao, S, Wang, W, Kuang, T, Sui, S, Shen, J. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-31 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex.
Science, 365, 2019
|
|
7EZX
 
 | |
6L35
 
 | | PSI-LHCI Supercomplex from Physcometrella patens | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Zhao, L, Yan, Q.J, Qin, X.C. | | Deposit date: | 2019-10-09 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens.
Cell Discov, 7, 2021
|
|
6LY5
 
 | | Organization and energy transfer in a huge diatom PSI-FCPI supercomplex | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Xiong, P, Caizhe, X. | | Deposit date: | 2020-02-13 | | Release date: | 2020-10-21 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural basis for energy transfer in a huge diatom PSI-FCPI supercomplex.
Nat Commun, 11, 2020
|
|
8J6Z
 
 | | Cryo-EM structure of the Arabidopsis thaliana photosystem I(PSI-LHCII-ST2) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7B
 
 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 2 (PSI-ST2) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
8J7A
 
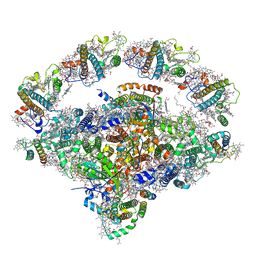 | | Coordinates of Cryo-EM structure of the Arabidopsis thaliana PSI in state 1 (PSI-ST1) | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
7DF8
 
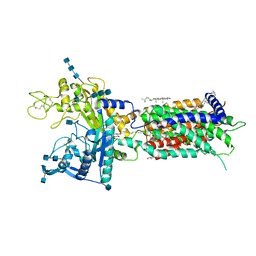 | | full length hNPC1L1-Apo | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, M, Sun, S, Sui, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake.
Sci Adv, 7, 2021
|
|
4IJS
 
 | |
6IGZ
 
 | | Structure of PSI-LHCI | | Descriptor: | (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Xiong, P, Xiaochun, Q. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structure of a green algal photosystem I in complex with a large number of light-harvesting complex I subunits.
Nat Plants, 5, 2019
|
|
