5XNM
 
 | | Structure of unstacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5XNN
 
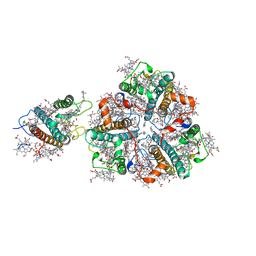 | | Structure of M-LHCII and CP24 complexes in the stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
7WFF
 
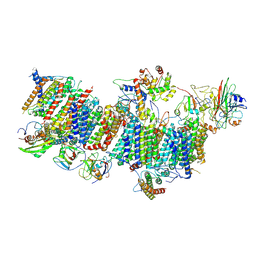 | | Subcomplexes B,M and L in the Cylic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WG5
 
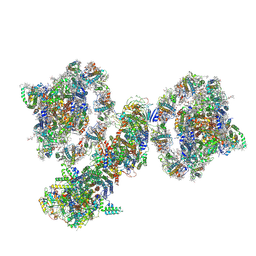 | | Cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-28 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFE
 
 | | Right PSI in the cyclic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFG
 
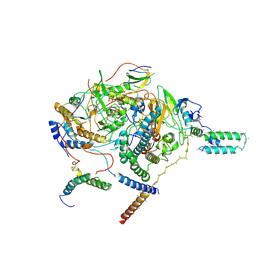 | | Subcomplexes A and E in NDH complex from Arabidopsis | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase subunit H, chloroplastic, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
3OEA
 
 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
3OEB
 
 | |
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
4RTH
 
 | | The crystal structure of PsbP from Zea mays | | Descriptor: | Membrane-extrinsic protein of photosystem II PsbP | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
4RTI
 
 | | The crystal structure of PsbP from Spinacia oleracea | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Oxygen-evolving enhancer protein 2, ... | | Authors: | Cao, P, Xie, Y, Li, M, Pan, X.W, Zhang, H.M, Zhao, X.L, Su, X.D, Cheng, T, Chang, W. | | Deposit date: | 2014-11-15 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of extrinsic PsbP protein of photosystem II reveals a manganese-induced conformational change.
Mol Plant, 8, 2015
|
|
5OFL
 
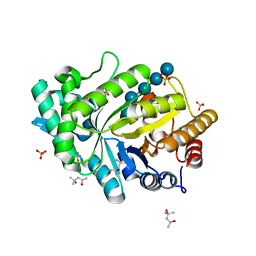 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, SULFATE ION, ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFK
 
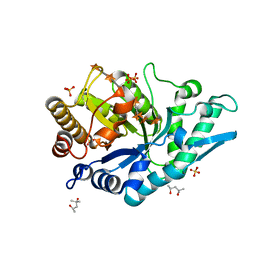 | | Crystal structure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glycoside hydrolase family 48, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), ... | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J, Tu, T. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
5OFJ
 
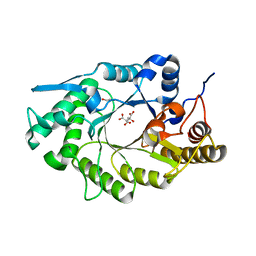 | | Crystal structure of N-terminal domain of bifunctional CbXyn10C | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Glycoside hydrolase family 48 | | Authors: | Hakulinen, N, Penttinen, L, Rouvinen, J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Insights into the roles of non-catalytic residues in the active site of a GH10 xylanase with activity on cellulose.
J. Biol. Chem., 292, 2017
|
|
2GEB
 
 | |
7VHR
 
 | | Apostichopus japonicus ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Wu, Y, Su, X.R, Ming, T.H. | | Deposit date: | 2021-09-22 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.756 Å) | | Cite: | Crystallographic characterization of a marine invertebrate ferritin from the sea cucumber Apostichopus japonicus.
Febs Open Bio, 12, 2022
|
|
6J0T
 
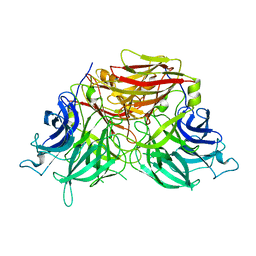 | | The crystal structure of exoinulinase INU1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Inulinase, MAGNESIUM ION | | Authors: | Hu, X.-J. | | Deposit date: | 2018-12-26 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure Study of Exoinulinase INU1
To Be Published
|
|
5TFP
 
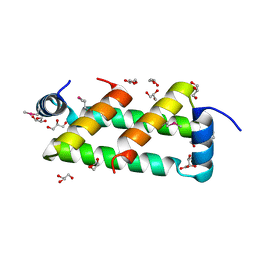 | |
5ZJI
 
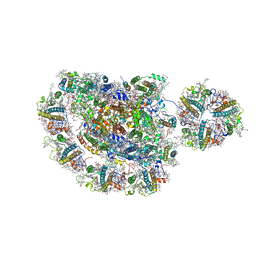 | | Structure of photosystem I supercomplex with light-harvesting complexes I and II | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Pan, X.W, Ma, J, Su, X.D, Cao, P, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2018-03-20 | | Release date: | 2018-06-20 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Science, 360, 2018
|
|
3KM8
 
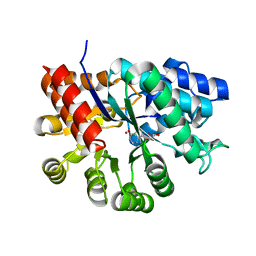 | |
3L8R
 
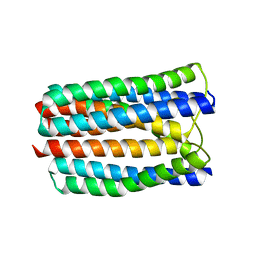 | | The crystal structure of PtcA from S. mutans | | Descriptor: | Putative PTS system, cellobiose-specific IIA component | | Authors: | Lei, J, Liu, X, Li, L. | | Deposit date: | 2010-01-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of PtcA from Streptococcus mutans
To be Published
|
|
8HCT
 
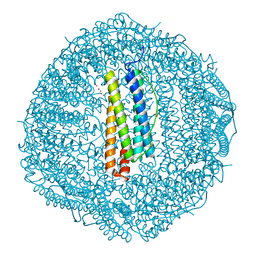 | | Crystal structure of Cu2+ binding to Dendrorhynchus zhejiangensis ferritin | | Descriptor: | COPPER (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Ming, T.H, Su, X.R, Huo, C.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Biochemical Characterization of Silver/Copper Binding by Dendrorhynchus zhejiangensis Ferritin.
Polymers (Basel), 15, 2023
|
|
8GY1
 
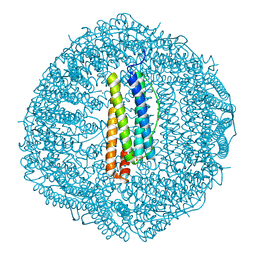 | |
6LPD
 
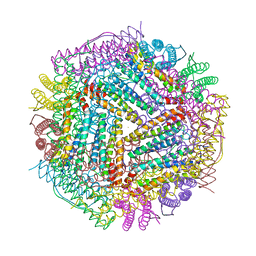 | | Phascolosoma esculenta | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin | | Authors: | Su, X.R, Ming, T.H. | | Deposit date: | 2020-01-09 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural comparison of two ferritins from the marine invertebrate Phascolosoma esculenta.
Febs Open Bio, 11, 2021
|
|
6LPE
 
 | | Phascolosoma esculenta ferritin | | Descriptor: | Ferritin, MAGNESIUM ION, SULFATE ION | | Authors: | Su, X.R, Ming, T.H. | | Deposit date: | 2020-01-10 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of two ferritins from the marine invertebrate Phascolosoma esculenta.
Febs Open Bio, 11, 2021
|
|
