7WAL
 
 | | Glutamyl-tRNA synthetase from Plasmodium falciparum (PfERS) in complex with Co | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Sharma, V, Manickam, Y, Babbar, P, Sharma, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Mol.Biochem.Parasitol., 253, 2022
|
|
7WAK
 
 | | Glutamyl-tRNA synthetase from Plasmodium falciparum (PfERS) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Glutamyl-tRNA synthetase, ... | | Authors: | Sharma, V, Manickam, Y, Babbar, P, Sharma, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.782 Å) | | Cite: | Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Mol.Biochem.Parasitol., 253, 2022
|
|
7WAJ
 
 | | Glutamyl-tRNA synthetase from Plasmodium falciparum (PfERS) complexed with ATP and Co | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Sharma, V, Manickam, Y, Babbar, P, Sharma, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Mol.Biochem.Parasitol., 253, 2022
|
|
7WAI
 
 | | Glutamyl-tRNA synthetase from Plasmodium falciparum (PfERS) | | Descriptor: | CHLORIDE ION, Glutamyl-tRNA synthetase | | Authors: | Sharma, V, Manickam, Y, Harlos, K, Sharma, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Mol.Biochem.Parasitol., 253, 2022
|
|
7WAO
 
 | | Glutamyl-tRNA synthetase from Plasmodium falciparum (PfERS) in complex with Mn | | Descriptor: | CHLORIDE ION, Glutamyl-tRNA synthetase, MANGANESE (II) ION | | Authors: | Sharma, V, Manickam, Y, Babbar, P, Sharma, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Structural characterization of glutamyl-tRNA synthetase (GluRS) from Plasmodium falciparum.
Mol.Biochem.Parasitol., 253, 2022
|
|
4R6O
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6P
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYL-2H-CHROMEN-2-ONE, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6N
 
 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | Descriptor: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | Authors: | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
8B6F
 
 | | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6H
 
 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6G
 
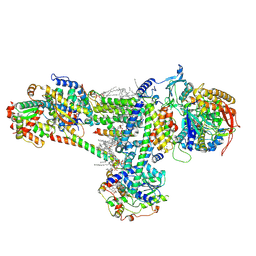 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6J
 
 | | Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apocytochrome b, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
4R6R
 
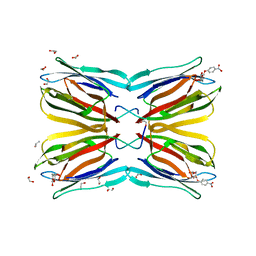 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-galactopyranoside, Agglutinin alpha chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R6Q
 
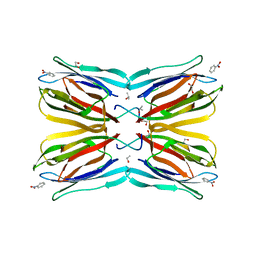 | | Jacalin-carbohydrate interactions. Distortion of the ligand as a determinant of affinity. | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin alpha chain, Agglutinin beta-3 chain, ... | | Authors: | Abhinav, K.V, Sharma, K, Swaminathan, C.P, Surolia, A, Vijayan, M. | | Deposit date: | 2014-08-26 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Jacalin-carbohydrate interactions: distortion of the ligand molecule as a determinant of affinity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4H02
 
 | |
7QRU
 
 | | Structure of Bacillus pseudofirmus Mrp antiporter complex, monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Na(+)/H(+) antiporter subunit B, Na(+)/H(+) antiporter subunit C, ... | | Authors: | Lee, Y. | | Deposit date: | 2022-01-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Ion transfer mechanisms in Mrp-type antiporters from high resolution cryoEM and molecular dynamics simulations.
Nat Commun, 13, 2022
|
|
8J4X
 
 | |
8J4V
 
 | |
8J4W
 
 | |
8J4Y
 
 | |
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex
To Be Published
|
|
7O71
 
 | | Cryo-EM structure of a respiratory complex I | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | High-resolution structure and dynamics of mitochondrial complex I-Insights into the proton pumping mechanism.
Sci Adv, 7, 2021
|
|
7O6Y
 
 | | Cryo-EM structure of respiratory complex I under turnover | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2021-04-12 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | High-resolution structure and dynamics of mitochondrial complex I-Insights into the proton pumping mechanism.
Sci Adv, 7, 2021
|
|
6Y79
 
 | | Cryo-EM structure of a respiratory complex I F89A mutant | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Essential role of accessory subunit LYRM6 in the mechanism of mitochondrial complex I.
Nat Commun, 11, 2020
|
|
