4HE2
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
4HE1
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase Q32R mutant complex with fructose-6-phosphate and phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
4HE0
 
 | | Crystal structure of human muscle fructose-1,6-bisphosphatase | | Descriptor: | CHLORIDE ION, Fructose-1,6-bisphosphatase isozyme 2, MAGNESIUM ION, ... | | Authors: | Shi, R, Zhu, D.W, Lin, S.X. | | Deposit date: | 2012-10-03 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal Structures of Human Muscle Fructose-1,6-Bisphosphatase: Novel Quaternary States, Enhanced AMP Affinity, and Allosteric Signal Transmission Pathway.
Plos One, 8, 2013
|
|
4FVR
 
 | |
5K55
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
5L0A
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
8GRO
 
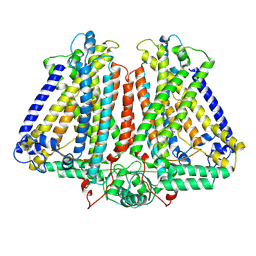 | | AtOSCA3.1 contracted state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GSO
 
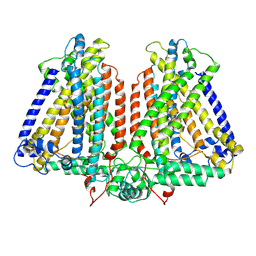 | | AtOSCA3.1 channel extended state | | Descriptor: | CSC1-like protein ERD4 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GRN
 
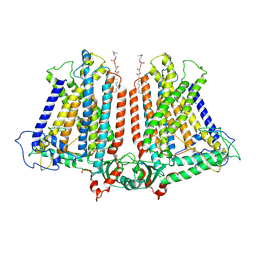 | | AtOSCA1.1 extended state | | Descriptor: | Protein OSCA1, [1-MYRISTOYL-GLYCEROL-3-YL]PHOSPHONYLCHOLINE | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
8GRS
 
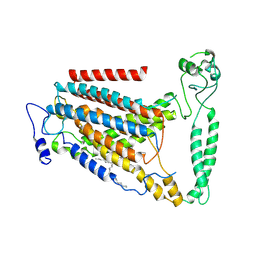 | | human TMEM63A | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CSC1-like protein 1 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-09-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A mechanical-coupling mechanism in OSCA/TMEM63 channel mechanosensitivity.
Nat Commun, 14, 2023
|
|
5ET5
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET8
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | T-to-R switch of muscle FBPase involves extreme changes of secondary and quaternary structure
To Be Published
|
|
5ET6
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ET7
 
 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6JXT
 
 | |
6JWL
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
6JX4
 
 | |
6JX0
 
 | |
6LTO
 
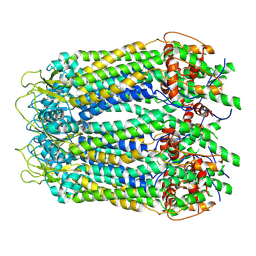 | | cryo-EM structure of full length human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
6LTN
 
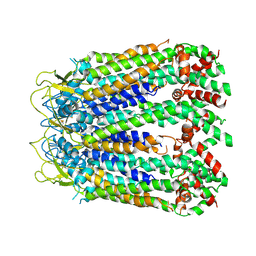 | | cryo-EM structure of C-terminal truncated human Pannexin1 | | Descriptor: | Pannexin-1 | | Authors: | Mou, L.Q, Ke, M, Xiao, Q.J, Wu, J.P, Deng, D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for gating mechanism of Pannexin 1 channel.
Cell Res., 30, 2020
|
|
7YID
 
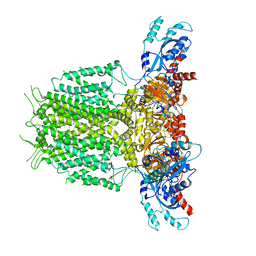 | | Human KCNH5 closed state 1 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
7YIE
 
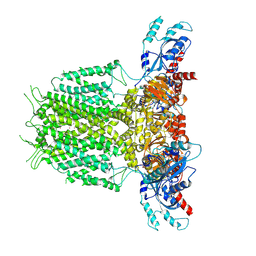 | | Human KCNH5-closed state 2 | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
7YIH
 
 | | Human KCNH5 open state | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily H member 5 | | Authors: | Zhang, M.F. | | Deposit date: | 2022-07-16 | | Release date: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism underlying delayed rectifying in human voltage-mediated activation Eag2 channel.
Nat Commun, 14, 2023
|
|
