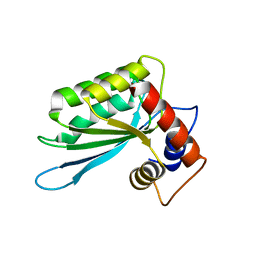
5F0Z
 
 | |
4DVD
 
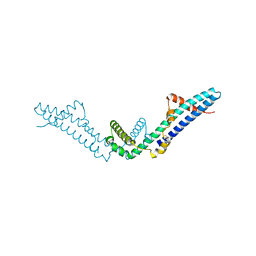 | |
5EY7
 
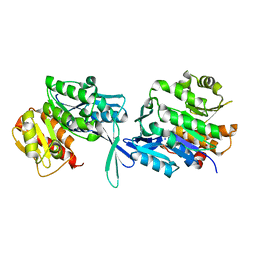 | |
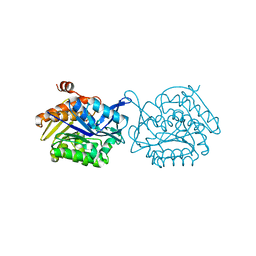
3IQS
 
 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
6K6T
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form | | Descriptor: | 9-[(1R,6R,8R,9S,10R,15S,17R,18S)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-17-(6-oxidanylidene-3H-purin-9-yl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-3H-purin-6-one, CALCIUM ION, EAL domain protein | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2019-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-IMP bound form
To Be Published
|
|
5JZM
 
 | |
5JZO
 
 | |
5K4A
 
 | |
6KRB
 
 | | High resolution crystal structure of an Acylphosphatase protein cage | | Descriptor: | Acylphosphatase, GLYCEROL, SULFATE ION | | Authors: | Chatterjee, S, Nath, S, Sen, U. | | Deposit date: | 2019-08-21 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.375 Å) | | Cite: | High resolution structure of Vibrio cholerae acylphosphatase (VcAcP) cage: Identification of drugs, location of its binding site and engineering to facilitate cage formation.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
3I29
 
 | | Crystal structure of a binary complex between an mutant trypsin inhibitor with bovine trypsin | | Descriptor: | CALCIUM ION, Cationic trypsin, Chymotrypsin inhibitor 3 | | Authors: | Khamrui, S, Majumder, S, Dasgupta, J, Dattagupta, J.K, Sen, U. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role of remote scaffolding residues in the inhibitory loop pre-organization, flexibility, rigidification and enzyme inhibition of serine protease inhibitors
Biochim.Biophys.Acta, 1824, 2012
|
|
6IH7
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - 3',3'-cGAMP bound form | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
6IH1
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-GMP bound form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
6IFQ
 
 | |
4X8F
 
 | | Vibrio cholerae O395 Ribokinase in apo form | | Descriptor: | Ribokinase | | Authors: | Paul, R, Patra, M.D, Sen, U. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Apo and Ligand
Bound Vibrio choleraeRibokinase (Vc-RK):
Role of Monovalent Cation Induced Activation
and Structural Flexibility in Sugar
Phosphorylation
Adv Exp Med Biol., 842, 2015
|
|
6IJ2
 
 | |
3RX6
 
 | | Crystal structure of Polarity Suppression protein from Enterobacteria phage P4 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, MERCURY (II) ION, ... | | Authors: | Banerjee, R, Nath, S, Khamrui, S, Sen, R, Sen, U. | | Deposit date: | 2011-05-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | The first structure of polarity suppression protein, Psu from enterobacteria phage P4, reveals a novel fold and a knotted dimer
J.Biol.Chem., 287, 2012
|
|
4HNQ
 
 | |
4HI2
 
 | |
4HNS
 
 | | Crystal structure of activated CheY3 of Vibrio cholerae | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, MAGNESIUM ION | | Authors: | Biswas, M, Sen, U, Dasgupta, J. | | Deposit date: | 2012-10-21 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational Barrier of CheY3 and Inability of CheY4 to Bind FliM Control the Flagellar Motor Action in Vibrio cholerae.
Plos One, 8, 2013
|
|
4HI1
 
 | |
4HNR
 
 | |
4I2N
 
 | |
4JP1
 
 | | Mg2+ bound structure of Vibrio Cholerae CheY3 | | Descriptor: | Chemotaxis protein CheY, MAGNESIUM ION | | Authors: | Biswas, M, Dasgupta, J, Sen, U. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae
Plos One, 8, 2013
|
|
4H9W
 
 | | crystal structure of a METHIONINE mutant of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Majumder, S, Sen, U. | | Deposit date: | 2012-09-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved tryptophan (W91) at the barrel-lid junction modulates the packing and stability of Kunitz (STI) family of inhibitors.
Biochim.Biophys.Acta, 1854, 2015
|
|
7V4E
 
 | | Crystal Structure of VpsR display novel dimeric architecture and c-di-GMP binding: mechanistic implications in oligomerization, ATPase activity and DNA binding. | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, VpsR | | Authors: | Chakrabortty, T, Sen, U. | | Deposit date: | 2021-08-12 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of VpsR Revealed Novel Dimeric Architecture and c-di-GMP Binding Site: Mechanistic Implications in Oligomerization, ATPase Activity and DNA Binding.
J.Mol.Biol., 434, 2022
|
|
