7TMF
 
 | |
7TMG
 
 | |
8FF9
 
 | | Crystal structure of Apo Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | CHLORIDE ION, Probable dna-binding stress protein, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8DOR
 
 | |
8FFA
 
 | |
8FFB
 
 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (orthorhombic form) | | Descriptor: | FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8FFC
 
 | | Crystal structure of iron bound Dps protein (PA0962) from Pseudomonas aeruginosa (cubic form) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE (II) ION, Probable dna-binding stress protein | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rivera, M. | | Deposit date: | 2022-12-08 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pseudomonas aeruginosa Dps (PA0962) Functions in H 2 O 2 Mediated Oxidative Stress Defense and Exhibits In Vitro DNA Cleaving Activity.
Int J Mol Sci, 24, 2023
|
|
8SBO
 
 | |
8SLF
 
 | |
8SLD
 
 | |
8SLG
 
 | |
8SLH
 
 | |
8T1A
 
 | |
8DOS
 
 | |
8DQC
 
 | |
8DOP
 
 | |
8DQB
 
 | |
8DOQ
 
 | |
8DQ9
 
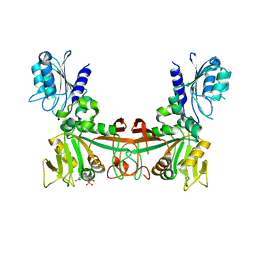 | |
8EES
 
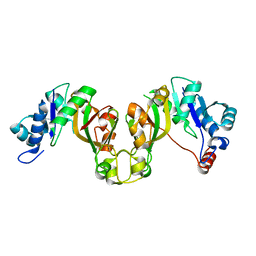 | |
8EK7
 
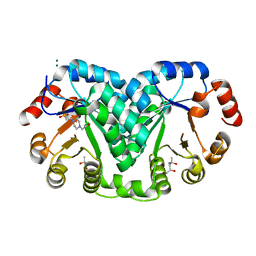 | |
8W1D
 
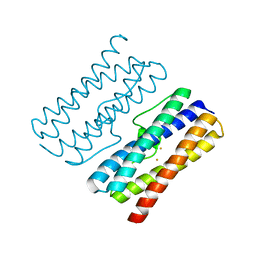 | | CRYSTAL STRUCTURE OF DPS-LIKE PROTEIN PA4880 FROM PSEUDOMONAS AERUGINOSA (DIMERIC FORM) | | Descriptor: | DPS-LIKE PROTEIN, FE (II) ION | | Authors: | Lovell, S, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-02-15 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pseudomonas aeruginosa gene PA4880 encodes a Dps-like protein with a Dps fold, bacterioferritin-type ferroxidase centers, and endonuclease activity.
Front Mol Biosci, 11, 2024
|
|
8TA0
 
 | |
8TBR
 
 | |
8TA1
 
 | |
