6Q2F
 
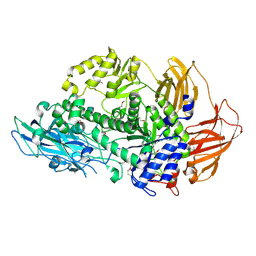 | | Structure of Rhamnosidase from Novosphingobium sp. PP1Y | | Descriptor: | Glycoside hydrolase family protein, SODIUM ION | | Authors: | Terry, B, Ha, J, Izzo, V, Sazinsky, M.H. | | Deposit date: | 2019-08-07 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.20000076 Å) | | Cite: | The crystal structure and insight into the substrate specificity of the alpha-L rhamnosidase RHA-P from Novosphingobium sp. PP1Y.
Arch.Biochem.Biophys., 679, 2019
|
|
4FX9
 
 | | Structure of the Pyrococcus horikoshii CoA persulfide/polysulfide reductase | | Descriptor: | COENZYME A, Coenzyme A disulfide reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Herwald, S, Lopez, K.M, Crane III, E.J, Sazinsky, M.H. | | Deposit date: | 2012-07-02 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and substrate specificity of the pyrococcal coenzyme A disulfide reductases/polysulfide reductases (CoADR/Psr): implications for S(0)-based respiration and a sulfur-dependent antioxidant system in Pyrococcus.
Biochemistry, 52, 2013
|
|
3FDC
 
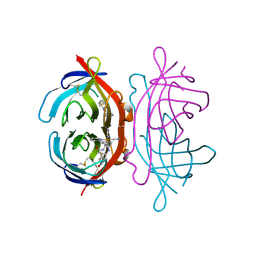 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
1FZ7
 
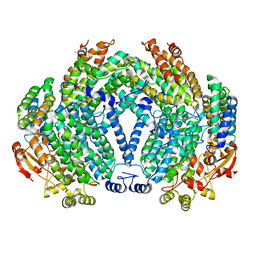 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM III SOAKED IN 0.9 M ETHANOL | | Descriptor: | CALCIUM ION, ETHANOL, FE (III) ION, ... | | Authors: | Whittington, D.A, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase.
J.Am.Chem.Soc., 123, 2001
|
|
3N1Y
 
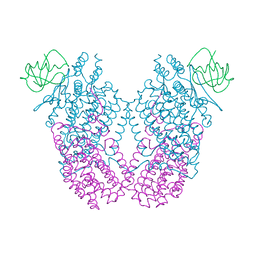 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase T201G Mutant | | Descriptor: | FE (III) ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Active Site Threonine Facilitates Proton Transfer during Dioxygen Activation at the Diiron Center of Toluene/o-Xylene Monooxygenase Hydroxylase.
J.Am.Chem.Soc., 132, 2010
|
|
3N1Z
 
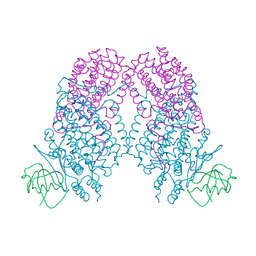 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase T201S Mutant | | Descriptor: | FE (III) ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Active Site Threonine Facilitates Proton Transfer during Dioxygen Activation at the Diiron Center of Toluene/o-Xylene Monooxygenase Hydroxylase.
J.Am.Chem.Soc., 132, 2010
|
|
1FZ6
 
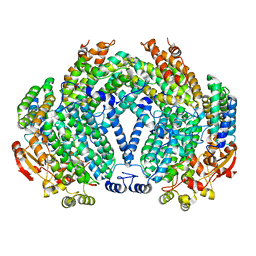 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM II SOAKED IN 1 M METHANOL | | Descriptor: | CALCIUM ION, FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ... | | Authors: | Whittington, D.A, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase.
J.Am.Chem.Soc., 123, 2001
|
|
3N20
 
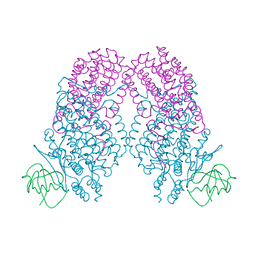 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase T201V Mutant | | Descriptor: | FE (III) ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active Site Threonine Facilitates Proton Transfer during Dioxygen Activation at the Diiron Center of Toluene/o-Xylene Monooxygenase Hydroxylase.
J.Am.Chem.Soc., 132, 2010
|
|
3RNB
 
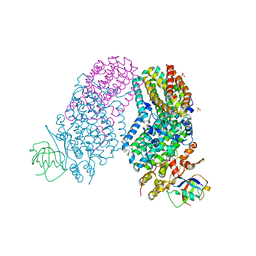 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/F176W Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, SULFATE ION, ... | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RNG
 
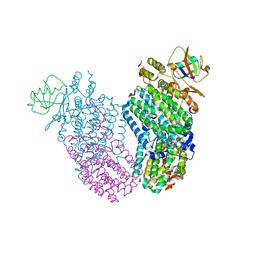 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/W167E Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Toluene o-xylene monooxygenase component | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RNA
 
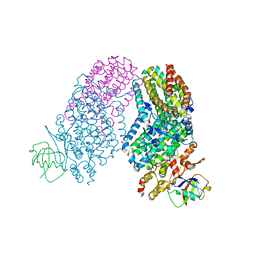 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/I100W Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Toluene o-xylene monooxygenase component | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RNC
 
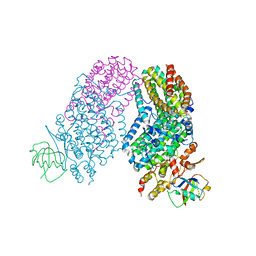 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/I100A Double Mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RNF
 
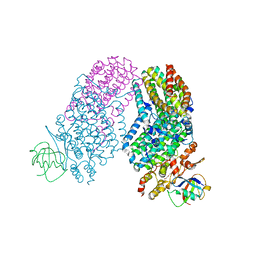 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/V271A Double Mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RNE
 
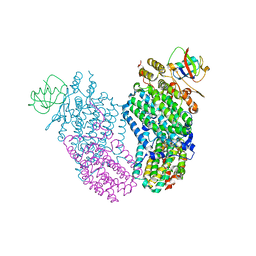 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/I276E Double Mutant | | Descriptor: | FE (III) ION, HYDROXIDE ION, Toluene o-xylene monooxygenase component | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RN9
 
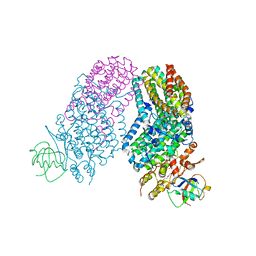 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase T201S/L272E Double Mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Gucinski, G, Song, W.J, Lippard, S.J, Sazinsky, M.H. | | Deposit date: | 2011-04-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tracking a defined route for O2 migration in a dioxygen-activating diiron enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IND
 
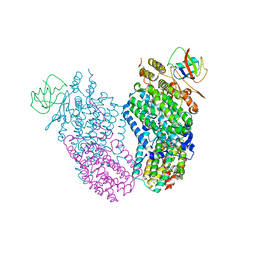 | | Mn(II) Reconstituted Toluene/o-xylene Monooxygenase Hydroxylase X-ray Crystal Structure | | Descriptor: | HEXAETHYLENE GLYCOL, MANGANESE (II) ION, Toluene, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Condon, K.L, Lippard, S.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
J.Am.Chem.Soc., 128, 2006
|
|
2INC
 
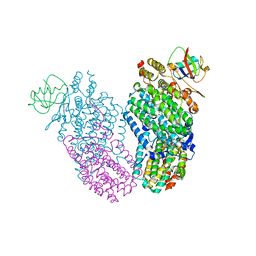 | | Native Toluene/o-xylene Monooxygenase Hydroxylase X-ray Crystal Structure | | Descriptor: | FE (III) ION, HEXAETHYLENE GLYCOL, Toluene, ... | | Authors: | McCormick, M.S, Sazinsky, M.H, Condon, K.L, Lippard, S.J. | | Deposit date: | 2006-10-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
J.Am.Chem.Soc., 128, 2006
|
|
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NTA
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NT6
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C43S/C531S Double Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H, Lopez, K. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
2INP
 
 | | Structure of the Phenol Hydroxylase-Regulatory Protein Complex | | Descriptor: | FE (III) ION, Phenol hydroxylase component phL, Phenol hydroxylase component phM, ... | | Authors: | Sazinsky, M.S, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
3FRY
 
 | | Crystal structure of the CopA C-terminal metal binding domain | | Descriptor: | CITRIC ACID, Probable copper-exporting P-type ATPase A | | Authors: | Agarwal, S, Sazinsky, M, Arguello, J, Rosenzweig, A.C. | | Deposit date: | 2009-01-08 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and interactions of the C-terminal metal binding domain of Archaeoglobus fulgidus CopA.
Proteins, 78, 2010
|
|
