3LIV
 
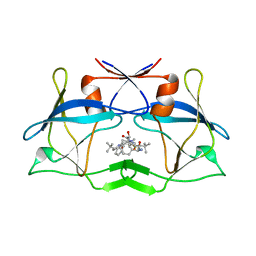 | | crystal structure of HTLV protease complexed with the inhibitor KNI-10683 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-N-[(2R)-3,3-dimethylbutan-2-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
3LIT
 
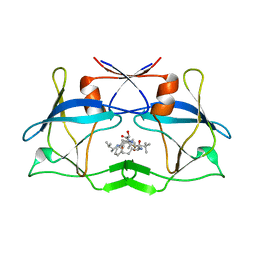 | | The crystal structure of htlv protease complexed with the inhibitor KNI-10681 | | Descriptor: | (4R)-3-[(2S,3S)-3-[[(2S)-2-[[(2S)-2-azanyl-2-phenyl-ethanoyl]amino]-3,3-dimethyl-butanoyl]amino]-2-hydroxy-4-phenyl-but anoyl]-5,5-dimethyl-N-[(2R)-3-methylbutan-2-yl]-1,3-thiazolidine-4-carboxamide, Protease | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
2A70
 
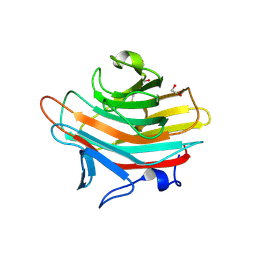 | | Crystal structure of Emp47p carbohydrate recognition domain (CRD), monoclinic crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, Emp47p | | Authors: | Satoh, T, Sato, K, Kanoh, A, Yamashita, K, Katoh, R, Nakano, A, Wakatsuki, S. | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structures of the carbohydrate recognition domain of Ca2+-independent cargo receptors Emp46p and Emp47p.
J.Biol.Chem., 281, 2006
|
|
3WHJ
 
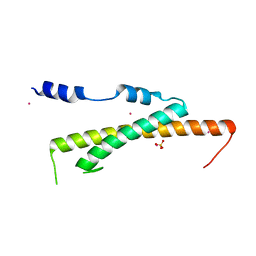 | | Crystal structure of Nas2 N-terminal domain | | Descriptor: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WNX
 
 | | Crystal structure of ERGIC-53/MCFD2, Calcium/Man3-bound form | | Descriptor: | CALCIUM ION, Multiple coagulation factor deficiency protein 2, Protein ERGIC-53, ... | | Authors: | Satoh, T, Suzuki, K, Yamaguchi, T, Kato, K. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis for Disparate Sugar-Binding Specificities in the Homologous Cargo Receptors ERGIC-53 and VIP36
Plos One, 9, 2014
|
|
2DUO
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36 | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUQ
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Vesicular integral-membrane protein VIP36, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
3WHU
 
 | | Crystal structure of ERGIC-53/MCFD2, Calcium/Man2-bound form | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Suzuki, K, Kato, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Disparate Sugar-Binding Specificities in the Homologous Cargo Receptors ERGIC-53 and VIP36
Plos One, 9, 2014
|
|
2E6V
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man3GlcNAc-bound form | | Descriptor: | CALCIUM ION, Vesicular integral-membrane protein VIP36, alpha-D-mannopyranose, ... | | Authors: | Satoh, T, Kato, R, Wakatsuki, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
3WHT
 
 | | Crystal structure of ERGIC-53/MCFD2, Calcium-free form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Suzuki, K, Kato, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Disparate Sugar-Binding Specificities in the Homologous Cargo Receptors ERGIC-53 and VIP36
Plos One, 9, 2014
|
|
2DUR
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, Ca2+/Man2-bound form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
2DUP
 
 | | Crystal structure of VIP36 exoplasmic/lumenal domain, metal-free form | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Satoh, T, Cowieson, N.P, Kato, R, Wakatsuki, S. | | Deposit date: | 2006-07-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of high mannose type glycoproteins by mammalian transport lectin VIP36
J.Biol.Chem., 282, 2007
|
|
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3AIH
 
 | | Human OS-9 MRH domain complexed with alpha3,alpha6-Man5 | | Descriptor: | Protein OS-9, alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose | | Authors: | Satoh, T, Chen, Y, Hu, D, Hanashima, S, Yamamoto, K, Yamaguchi, Y. | | Deposit date: | 2010-05-14 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Oligosaccharide Recognition of Misfolded Glycoproteins by OS-9 in ER-Associated Degradation
Mol.Cell, 40, 2010
|
|
5DSV
 
 | | Crystal structure of human proteasome alpha7 tetradecamer | | Descriptor: | Proteasome subunit alpha type-3 | | Authors: | Satoh, T, Thammaporn, R, Seetaha, S, Kato, K. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Disassembly of the self-assembled, double-ring structure of proteasome alpha 7 homo-tetradecamer by alpha 6
Sci Rep, 5, 2015
|
|
1WNI
 
 | | Crystal Structure of H2-Proteinase | | Descriptor: | Trimerelysin II, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | Deposit date: | 2004-08-04 | | Release date: | 2004-08-17 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
5BW7
 
 | | Crystal structure of nonfucosylated Fc Y296W mutant complexed with bis-glycosylated soluble form of Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Isoda, Y, Yagi, H, Satoh, T, Shibata-Koyama, M, Masuda, K, Satoh, M, Kato, K, Iida, S. | | Deposit date: | 2015-06-06 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Importance of the Side Chain at Position 296 of Antibody Fc in Interactions with Fc gamma RIIIa and Other Fc gamma Receptors
Plos One, 10, 2015
|
|
8IC9
 
 | | Lys48-linked K48C-diubiquitin | | Descriptor: | Polyubiquitin-B, Ubiquitin | | Authors: | Hiranyakorn, M, Yagi-Utsumi, M, Yanaka, S, Ohtsuka, N, Momiyama, N, Satoh, T, Kato, K. | | Deposit date: | 2023-02-11 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mutational and Environmental Effects on the Dynamic Conformational Distributions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 24, 2023
|
|
7BPG
 
 | | Structure of serinol nucleic acid - RNA complex | | Descriptor: | CALCIUM ION, RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3'), SNA (S-(F7R)(F7X)(F7O)(F7R)(F7X)(F7O)(F7R)(F7U)-R) | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
7BPF
 
 | | Structure of L-threoninol nucleic acid - RNA complex | | Descriptor: | L-aTNA (3'-(*GP*CP*AP*GP*CP*AP*GP*C)-1'), RNA (5'-R(*GP*CP*UP*GP*CP*(5BU)P*GP*C)-3') | | Authors: | Kamiya, Y, Satoh, T, Kodama, A, Suzuki, T, Uchiyama, S, Kato, K, Asanuma, H. | | Deposit date: | 2020-03-22 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Intrastrand backbone-nucleobase interactions stabilize unwound right-handed helical structures of heteroduplexes of L-aTNA/RNA and SNA/RNA
Commun Chem, 2020
|
|
6KRU
 
 | | Crystal structure of mouse IgG2b Fc | | Descriptor: | Ig gamma-2B chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
6KRV
 
 | | Crystal structure of mouse IgG2b Fc complexed with B domain of Protein A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2B chain C region, ... | | Authors: | Taniguchi, Y, Satoh, T, Yagi, H, Kato, K. | | Deposit date: | 2019-08-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | On-Membrane Dynamic Interplay between Anti-GM1 IgG Antibodies and Complement Component C1q.
Int J Mol Sci, 21, 2019
|
|
2RUF
 
 | |
