8RPR
 
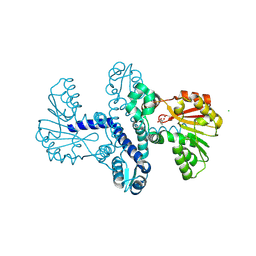 | | Crystal Structure of SgvM methyltransferase in complex with alpha-ketoleucine and Zn2+ ion | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Saleem-Batcha, R, Zou, Z, Breiltgens, J, Mueller, M, Andexer, J.N. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 25, 2024
|
|
6SGN
 
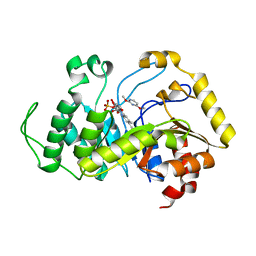 | | Crystal structure of monooxygenase RutA complexed with 2,4-dimethoxypyrimidine. | | Descriptor: | 2,4-dimethoxypyrimidine, FLAVIN MONONUCLEOTIDE, Pyrimidine monooxygenase RutA | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SGG
 
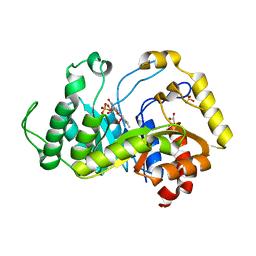 | | Crystal structure of monooxygenase RutA complexed with dioxygen under 1.5 MPa / 15 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-04 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SGL
 
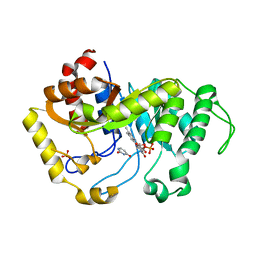 | | Crystal structure of monooxygenase RutA complexed with Uracil under atmospheric pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Pyrimidine monooxygenase RutA, SULFATE ION, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SLB
 
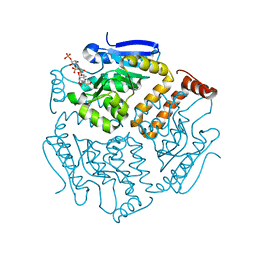 | | Crystal structure of isomerase PaaG with trans-3,4-didehydroadipyl-CoA | | Descriptor: | (~{E})-6-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethylsulfanyl]-6-oxidanylidene-hex-3-enoic acid, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
6SL9
 
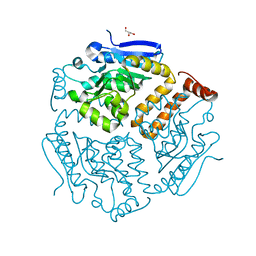 | |
6SLA
 
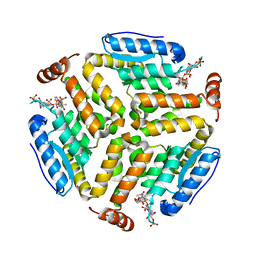 | | Crystal structure of isomerase PaaG mutant - D136N with Oxepin-CoA | | Descriptor: | Enoyl-CoA hydratase/carnithine racemase, ~{S}-[2-[3-[[(2~{R})-4-[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 2-(2,5-dihydrooxepin-7-yl)ethanethioate | | Authors: | Saleem-Batcha, R, Spieker, M, Teufel, R. | | Deposit date: | 2019-08-19 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Mechanistic Basis of an Oxepin-CoA Forming Isomerase in Bacterial Primary and Secondary Metabolism.
Acs Chem.Biol., 14, 2019
|
|
6SGM
 
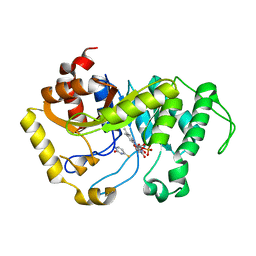 | |
6TEE
 
 | |
6TEG
 
 | | Crystal structure of monooxygenase RutA complexed with uracil and dioxygen under 1.5 MPa / 15 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN MOLECULE, Pyrimidine monooxygenase RutA, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-11-11 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6Q40
 
 | | A secreted LysM effector of the wheat pathogen Zymoseptoria tritici protects the fungal hyphae against chitinase hydrolysis through ligand-dependent polymerisation of LysM homodimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LysM domain-containing protein | | Authors: | Mesters, J.R, Saleem-Batcha, R, Sanchez-Vallet, A, Thomma, B.P.H.J. | | Deposit date: | 2018-12-05 | | Release date: | 2019-10-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization.
Plos Pathog., 16, 2020
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
8R4Z
 
 | |
8RVC
 
 | | Crystal structure of alpha keto acid C-methyl-transferases MrsA bound to ketoarginine | | Descriptor: | 1,2-ETHANEDIOL, 2-ketoarginine methyltransferase, 5-[(diaminomethylidene)amino]-2-oxopentanoic acid, ... | | Authors: | Gerhardt, S, Kemper, F, Andexer, J.N. | | Deposit date: | 2024-02-01 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structures and Protein Engineering of the alpha-Keto Acid C-Methyltransferases SgvM and MrsA for Rational Substrate Transfer.
Chembiochem, 25, 2024
|
|
8RWM
 
 | |
8RWW
 
 | |
8RXG
 
 | |
8RVS
 
 | |
8RXF
 
 | |
9H8L
 
 | | Crystal Structure of Polyphosphate kinase 2-II (PPK2-II) from Lysinibacillus fusiformis bound to TMP | | Descriptor: | PHOSPHATE ION, Polyphosphate kinase, THYMIDINE-5'-PHOSPHATE | | Authors: | Friedrich, F, Kuge, M, Keppler, M, Gerhardt, S, Einsle, O, Andexer, J.N. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Broad-Range Polyphosphate Kinase 2-II Enzymes Applicable for Pyrimidine Nucleoside Diphosphate Synthesis.
Chembiochem, 26, 2025
|
|
9H8J
 
 | | Crystal Structure of Polyphosphate kinase 2-II (PPK2-II) from Lysinibacillus fusiformis in apo form | | Descriptor: | PHOSPHATE ION, Polyphosphate kinase, SUCCINIC ACID | | Authors: | Friedrich, F, Kuge, M, Keppler, M, Gerhardt, S, Einsle, O, Andexer, J.N. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into Broad-Range Polyphosphate Kinase 2-II Enzymes Applicable for Pyrimidine Nucleoside Diphosphate Synthesis.
Chembiochem, 26, 2025
|
|
9H8K
 
 | | Crystal Structure of Polyphosphate kinase 2-II (PPK2-II) from Lysinibacillus fusiformis bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Friedrich, F, Kuge, M, Keppler, M, Gerhardt, S, Einsle, O, Andexer, J.N. | | Deposit date: | 2024-10-29 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Insights into Broad-Range Polyphosphate Kinase 2-II Enzymes Applicable for Pyrimidine Nucleoside Diphosphate Synthesis.
Chembiochem, 26, 2025
|
|
