6GGS
 
 | | Structure of RIP2 CARD filament | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pellegrini, E, Cusack, S, Desfosses, A, Schoehn, G, Malet, H, Gutsche, I, Sachse, C, Hons, M. | | Deposit date: | 2018-05-03 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | RIP2 filament formation is required for NOD2 dependent NF-kappa B signalling.
Nat Commun, 9, 2018
|
|
6TGN
 
 | |
6TGP
 
 | |
6TGY
 
 | |
6TH3
 
 | |
5AEY
 
 | | actin-like ParM protein bound to AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles.
Nature, 523, 2015
|
|
6SAE
 
 | | Cryo-EM structure of TMV in water | | Descriptor: | Capsid protein, MAGNESIUM ION, RNA (5'-R(P*GP*AP*A)-3') | | Authors: | Weis, F, Beckers, M, Sachse, C. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Elucidation of the viral disassembly switch of tobacco mosaic virus.
Embo Rep., 20, 2019
|
|
6SAG
 
 | | Cryo-EM structure of TMV with Ca2+ at low pH | | Descriptor: | CALCIUM ION, Capsid protein, MAGNESIUM ION, ... | | Authors: | Weis, F, Beckers, M, Sachse, C. | | Deposit date: | 2019-07-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Elucidation of the viral disassembly switch of tobacco mosaic virus.
Embo Rep., 20, 2019
|
|
5FJ8
 
 | | Cryo-EM structure of yeast RNA polymerase III elongation complex at 3. 9 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJ9
 
 | | Cryo-EM structure of yeast apo RNA polymerase III at 4.6 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
5FJA
 
 | | Cryo-EM structure of yeast RNA polymerase III at 4.7 A | | Descriptor: | DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC1, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC10, DNA-DIRECTED RNA POLYMERASE III SUBUNIT RPC2, ... | | Authors: | Hoffmann, N.A, Jakobi, A.J, Moreno-Morcillo, M, Glatt, S, Kosinski, J, Hagen, W.J, Sachse, C, Muller, C.W. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.65 Å) | | Cite: | Molecular Structures of Unbound and Transcribing RNA Polymerase III.
Nature, 528, 2015
|
|
6TGS
 
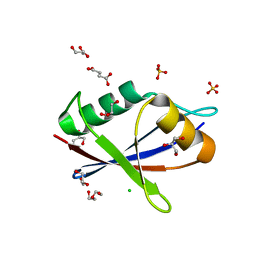 | | AtNBR1-PB1 domain | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Jakobi, A.J, Sachse, C. | | Deposit date: | 2019-11-17 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake.
Nat Commun, 11, 2020
|
|
9EM8
 
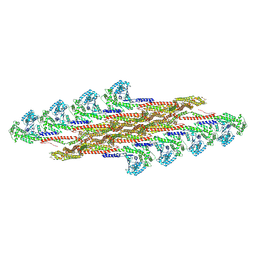 | | Oligomeric structure of SynDLP in presence of GDP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
9EM9
 
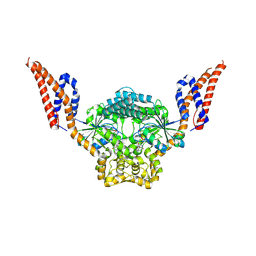 | | Structure of SynDLP MGD with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
9EM7
 
 | | Oligomeric structure of SynDLP in presence of GTP | | Descriptor: | Slr0869 protein | | Authors: | Junglas, B, Gewehr, L, Schoennenbeck, P, Schneider, D, Sachse, C. | | Deposit date: | 2024-03-07 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for GTPase activity and conformational changes of the bacterial dynamin-like protein SynDLP.
Cell Rep, 43, 2024
|
|
8QFV
 
 | | 305A Vipp1 helical tubes in the presence of EPL | | Descriptor: | Protein sll0617 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-05 | | Release date: | 2024-09-11 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QHX
 
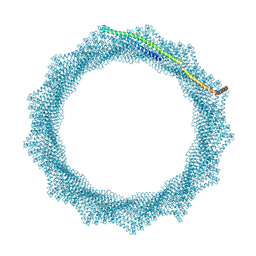 | | 280A Vipp1 H1-6 helical tubes in the presence of EPL | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QI0
 
 | | 340A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QI4
 
 | | 400A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QHY
 
 | | 300A Vipp1 H1-6 helical tubes in the presence of EPL | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QI5
 
 | | 405A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QHV
 
 | | 275A Vipp1 helical tubes in the presence of EPL | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QI2
 
 | | 370A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QI1
 
 | | 360A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QHZ
 
 | | 320A Vipp1 H1-6 helical tubes | | Descriptor: | Membrane-associated protein Vipp1 | | Authors: | Junglas, B, Sachse, C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-09-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structural basis for Vipp1 membrane binding: from loose coats and carpets to ring and rod assemblies.
Nat.Struct.Mol.Biol., 32, 2025
|
|
