4JQH
 
 | |
6NBW
 
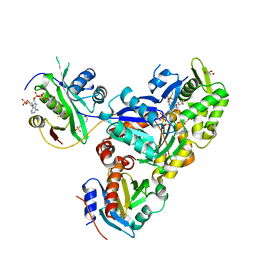 | | Ternary Complex of Beta/Gamma-Actin with Profilin and AnCoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-10 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NAS
 
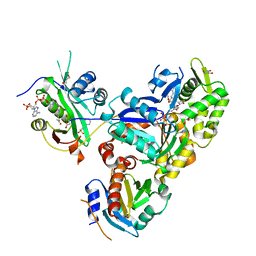 | | Ternary Complex of Ac-Alpha-Actin with Profilin and AcCoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-06 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
6NBE
 
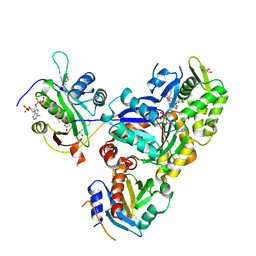 | | Ternary Complex of Ac-Alpha-Actin with Profilin and CoA-NAA80 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Rebowski, G, Boczkowska, M, Dominguez, R. | | Deposit date: | 2018-12-07 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of actin N-terminal acetylation.
Sci Adv, 6, 2020
|
|
5WJD
 
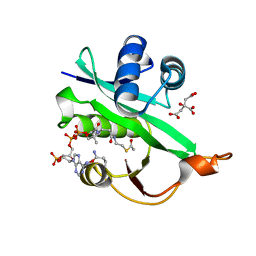 | | Crystal structure of Naa80 bound to acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, CG8481, isoform B, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WJE
 
 | | Crystal structure of Naa80 bound to a bisubstrate analogue | | Descriptor: | Actin N-terminus peptide, CARBOXYMETHYL COENZYME *A, CG8481, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5L7J
 
 | |
5L7L
 
 | |
6ZX9
 
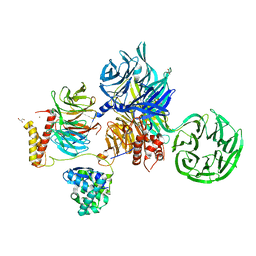 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
6ZUE
 
 | |
4JA9
 
 | | Rat PP5 apo | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
4JA7
 
 | | Rat PP5 co-crystallized with P5SA-2 | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
