8B12
 
 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-09 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
8B11
 
 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A and CsoS2 co-expression (T = 4) | | Descriptor: | Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
8B0Y
 
 | | cryo-EM structure of carboxysomal mini-shell: icosahedral assembly from CsoS4A/1A co-expression (T = 3) | | Descriptor: | Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Ni, T, Jiang, Q, Liu, L.N, Zhang, P. | | Deposit date: | 2022-09-08 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Intrinsically disordered CsoS2 acts as a general molecular thread for alpha-carboxysome shell assembly.
Nat Commun, 14, 2023
|
|
7ZC1
 
 | | Subtomogram averaging of Rubisco from Cyanobium carboxysome | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase, small subunit | | Authors: | Ni, T, Zhu, Y, Seaton-Burn, W, Zhang, P. | | Deposit date: | 2022-03-25 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and assembly of cargo Rubisco in two native alpha-carboxysomes.
Nat Commun, 13, 2022
|
|
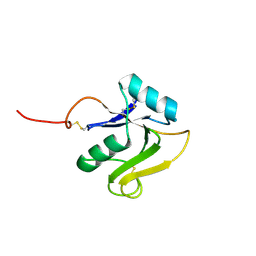
8BCM
 
 | |
8ORX
 
 | |
8OS2
 
 | | Structure of the human LYVE-1 (lymphatic vessel endothelial receptor-1) hyaluronan binding domain in an unliganded state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Lymphatic vessel endothelial hyaluronic acid receptor 1 | | Authors: | Ni, T, Bannerji, S, Jackson, D.J, Gilbert, R.J.C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A novel sliding interaction with hyaluronan underlies the mode of action of LYVE-1, the receptor for leucocyte entry and trafficking in the lymphatics
To be published
|
|
8YAX
 
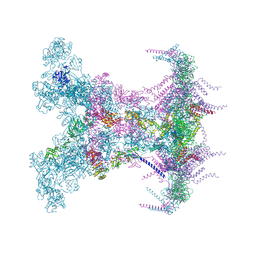 | | SARS-CoV-2 DMV nsp3-4 pore complex (full-pore) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-10 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB7
 
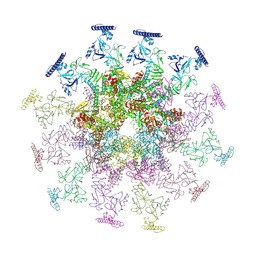 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C3 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
8YB5
 
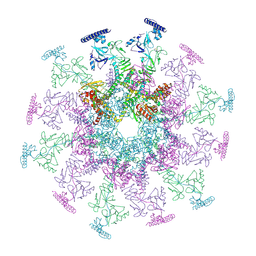 | | SARS-CoV-2 DMV nsp3-4 pore complex (consensus-pore, C6 symmetry) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
5L5N
 
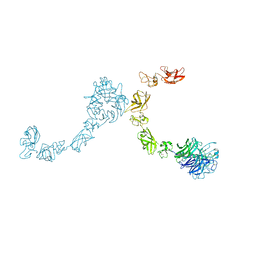 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8.5 angstrom, spacegroup P4(3)22 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (8.502 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5G
 
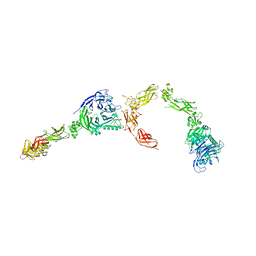 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5M
 
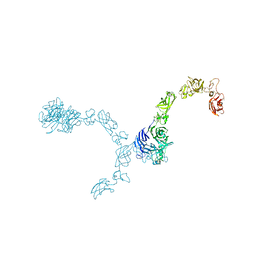 | | Plexin A4 full extracellular region, domains 1 to 7 modeled, data to 8 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L59
 
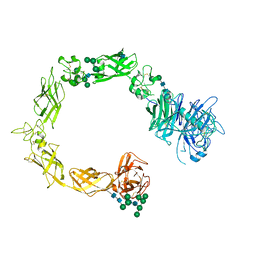 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P2(1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5K
 
 | | Plexin A4 full extracellular region, domains 1 to 10, data to 7.5 angstrom, spacegroup P4(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (7.501 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5L
 
 | | Plexin A4 full extracellular region, domains 1 to 8 modeled, data to 8 angstrom, spacegroup P2(1) | | Descriptor: | Plexin-A4 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L74
 
 | | Plexin A2 extracellular segment domains 4-5 (PSI2-IPT2), resolution 1.36 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Plexin-A2, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L7N
 
 | | Plexin A1 extracellular fragment, domains 7-10 (IPT3-IPT6) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kong, Y, Janssen, B.J.C, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L56
 
 | | Plexin A1 full extracellular region, domains 1 to 10, to 4 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L5C
 
 | | Plexin A1 full extracellular region, domains 1 to 10, to 6 angstrom, spacegroup P4(3)2(1)2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1, ... | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
7PLN
 
 | | Structure of the APCbeta domain of Plasmodium vivax perforin-like protein 1 | | Descriptor: | Sporozoite micronemal protein essential for cell traversal, putative | | Authors: | Williams, S.I, Ni, T, Yu, X, Gilbert, R.J.C. | | Deposit date: | 2021-08-31 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural, Functional and Computational Studies of Membrane Recognition by Plasmodium Perforin-Like Proteins 1 and 2
J.Mol.Biol., 434, 2022
|
|
6SNW
 
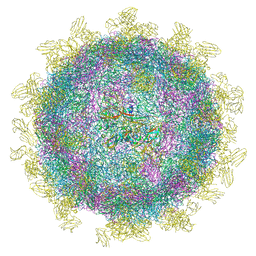 | | Structure of Coxsackievirus A10 complexed with its receptor KREMEN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6SMG
 
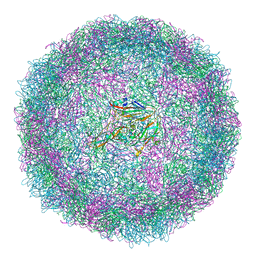 | | Structure of Coxsackievirus A10 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-21 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
6SNB
 
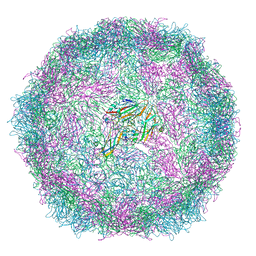 | | Structure of Coxsackievirus A10 A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-23 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
8A1S
 
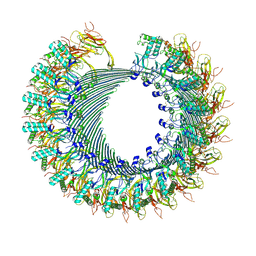 | | Structure of murine perforin-2 (Mpeg1) pore in twisted form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Yu, X, Ni, T, Zhang, P, Gilbert, R. | | Deposit date: | 2022-06-02 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structures of perforin-2 in isolation and assembled on a membrane suggest a mechanism for pore formation.
Embo J., 41, 2022
|
|
