2AO5
 
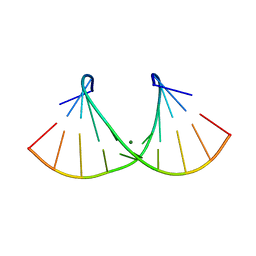 | | Crystal structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G-U base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*(5BU)P*GP*CP*GP*CP*U)-3', MAGNESIUM ION | | Authors: | Utsunomiya, R, Suto, K, Balasundaresan, D, Fukamizu, A, Kumar, P.K, Mizuno, H. | | Deposit date: | 2005-08-12 | | Release date: | 2006-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an RNA duplex r(GGCGBrUGCGCU)2 with terminal and internal tandem G.U base pairs.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1WRQ
 
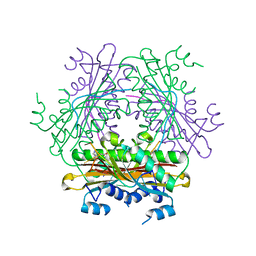 | |
1WRO
 
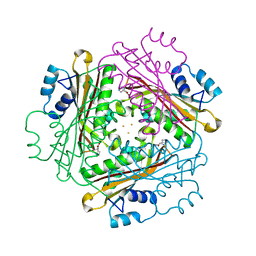 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | Descriptor: | BARIUM ION, HISTIDINE, Hut operon positive regulatory protein | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1WRN
 
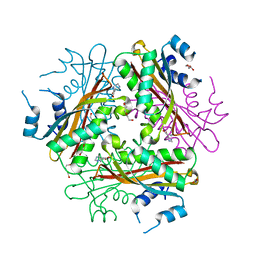 | | Metal Ion dependency of the antiterminator protein, HutP, for binding to the terminator region of hut mRNA- A structural basis | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-10-25 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1UEX
 
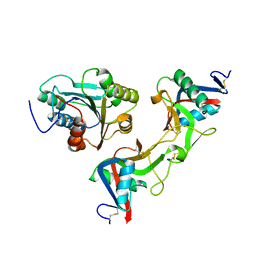 | | Crystal structure of von Willebrand Factor A1 domain complexed with snake venom bitiscetin | | Descriptor: | bitiscetin alpha chain, bitiscetin beta chain, von Willebrand Factor | | Authors: | Maita, N, Nishio, K, Nishimoto, E, Matsui, T, Shikamoto, Y, Morita, T, Sadler, J.E, Mizuno, H. | | Deposit date: | 2003-05-22 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of von Willebrand factor A1 domain complexed with snake venom, bitiscetin. Insight into glycoprotein Ibalpha binding mechanism induced by snake venom proteins.
J.Biol.Chem., 278, 2003
|
|
1J2L
 
 | | Crystal structure of the disintegrin, trimestatin | | Descriptor: | Disintegrin triflavin, SULFATE ION | | Authors: | Fujii, Y, Okuda, D, Fujimoto, Z, Morita, T, Mizuno, H. | | Deposit date: | 2003-01-06 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Trimestatin, a Disintegrin Containing a Cell Adhesion Recognition Motif RGD
J.Mol.Biol., 332, 2003
|
|
1HVX
 
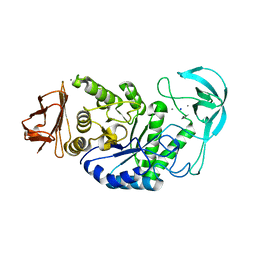 | | BACILLUS STEAROTHERMOPHILUS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Suvd, D, Fujimoto, Z, Takase, K, Matsumura, M, Mizuno, H. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability.
J.Biochem., 129, 2001
|
|
1WPT
 
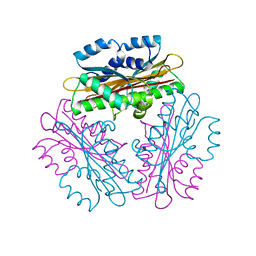 | | Crystal Structure of HutP, an RNA binding anti-termination protein | | Descriptor: | Hut operon positive regulatory protein | | Authors: | Kumarevel, T, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-13 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of the metal ion binding site in the anti-terminator protein, HutP, of Bacillus subtilis
Nucleic Acids Res., 33, 2005
|
|
1WMQ
 
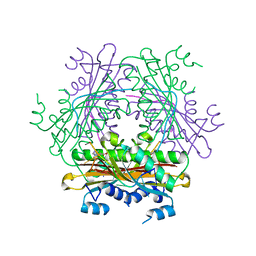 | | Structure of the HutP antitermination complex bound to a single stranded region of hut mRNA | | Descriptor: | 5'-R(P*UP*UP*UP*AP*GP*UP*U)-3', HISTIDINE, Hut operon positive regulatory protein, ... | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-07-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPV
 
 | | Crystal Structure of Activated Binary complex of HutP, an RNA binding anti-termination protein | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-09-14 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of HutP-mediated anti-termination and roles of the Mg2+ ion and L-histidine ligand.
Nature, 434, 2005
|
|
1WPU
 
 | |
1WPS
 
 | |
4H4L
 
 | | Crystal Structure of ternary complex of HutP(HutP-L-His-Zn) | | Descriptor: | HISTIDINE, Hut operon positive regulatory protein, ZINC ION | | Authors: | Dhakshnamoorthy, B, Misono, T.S, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alternative binding modes of l-histidine guided by metal ions for the activation of the antiterminator protein HutP of Bacillus subtilis.
J.Struct.Biol., 183, 2013
|
|
7X2B
 
 | | Red fluorescent protein from Diadumene lineata | | Descriptor: | Red fluorescent protein, SULFATE ION | | Authors: | Makabe, K, Hotta, J, Mizuno, H. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cloning and structural basis of fluorescent protein color variants from identical species of sea anemone, Diadumene lineata.
Photochem Photobiol Sci, 22, 2023
|
|
1XYF
 
 | | ENDO-1,4-BETA-XYLANASE FROM STREPTOMYCES OLIVACEOVIRIDIS | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Fujimoto, Z, Mizuno, H, Kuno, A, Kusakabe, I. | | Deposit date: | 1999-05-11 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Streptomyces olivaceoviridis E-86 beta-xylanase containing xylan-binding domain.
J.Mol.Biol., 300, 2000
|
|
2ZFA
 
 | | Structure of Lactate Oxidase at pH4.5 from AEROCOCCUS VIRIDANS | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Lactate oxidase | | Authors: | Furuichi, M, Balasundaresan, D, Suzuki, N, Yoshida, Y, Minagawa, H, Kaneko, H, Waga, I, Kumar, P.K.R, Mizuno, H. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray structures of Aerococcus viridans lactate oxidase and its complex with D-lactate at pH 4.5 show an alpha-hydroxyacid oxidation mechanism
J.Mol.Biol., 378, 2008
|
|
1UKM
 
 | | Crystal structure of EMS16, an Antagonist of collagen receptor integrin alpha2beta1 (GPIa/IIa) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of EMS16, an Antagonist of collagen receptor (GPIa/IIa) from the venom of Echis multisquamatus
Biochemistry, 42, 2003
|
|
2DD7
 
 | | A GFP-like protein from marine copepod, Chiridius poppei | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, green fluorescent protein | | Authors: | Suto, K, Masuda, H, Takenaka, Y, Mizuno, H. | | Deposit date: | 2006-01-23 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for red-shifted emission of a GFP-like protein from the marine copepod Chiridius poppei
Genes Cells, 14, 2009
|
|
2DD9
 
 | | A mutant of GFP-like protein from Chiridius poppei | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, green fluorescent protein | | Authors: | Suto, K, Masuda, H, Takenaka, Y, Mizuno, H. | | Deposit date: | 2006-01-24 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for red-shifted emission of a GFP-like protein from the marine copepod Chiridius poppei
Genes Cells, 14, 2009
|
|
1VEA
 
 | | Crystal Structure of HutP, an RNA binding antitermination protein | | Descriptor: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | Authors: | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
1V7P
 
 | | Structure of EMS16-alpha2-I domain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-12-19 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EMS16 in complex with the integrin alpha2-I domain
J.Mol.Biol., 341, 2004
|
|
2E1M
 
 | | Crystal Structure of L-Glutamate Oxidase from Streptomyces sp. X-119-6 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-glutamate oxidase, PHOSPHATE ION | | Authors: | Sasaki, C, Kashima, A, Sakaguchi, C, Mizuno, H, Arima, J, Kusakabe, H, Tamura, T, Sugio, S, Inagaki, K. | | Deposit date: | 2006-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of l-glutamate oxidase from Streptomyces sp. X-119-6
Febs J., 276, 2009
|
|
1VCK
 
 | | Crystal structure of ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10 | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, HYDROSULFURIC ACID, ... | | Authors: | Nam, J.-W, Noguchi, H, Fujiomoto, Z, Mizuno, H, Fushinobu, S, Kobashi, N, Iwata, K, Yoshida, T, Habe, H, Yamane, H, Omori, T, Nojiri, H. | | Deposit date: | 2004-03-09 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ferredoxin component of carbazole 1,9a-dioxygenase of Pseudomonas resinovorans strain CA10, a novel Rieske non-heme iron oxygenase system
PROTEINS, 58, 2005
|
|
1V6Y
 
 | | Crystal Structure Of chimeric Xylanase between Streptomyces Olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex | | Descriptor: | Beta-xylanase,Exoglucanase/xylanase | | Authors: | Kaneko, S, Ichinose, H, Fujimoto, Z, Kuno, A, Yura, K, Go, M, Mizuno, H, Kusakabe, I, Kobayashi, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of a family 10 beta-xylanase chimera of Streptomyces olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex
J.Biol.Chem., 279, 2004
|
|
1JWI
 
 | | Crystal Structure of Bitiscetin, a von Willeband Factor-dependent Platelet Aggregation Inducer. | | Descriptor: | bitiscetin, platelet aggregation inducer | | Authors: | Hirotsu, S, Mizuno, H, Fukuda, K, Qi, M.C, Matsui, T, Hamako, J, Morita, T, Titani, K. | | Deposit date: | 2001-09-04 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of bitiscetin, a von Willebrand factor-dependent platelet aggregation inducer.
Biochemistry, 40, 2001
|
|
