6YNU
 
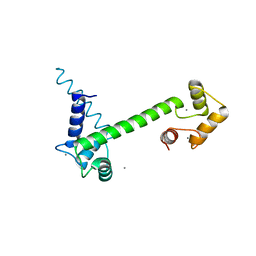 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YNS
 
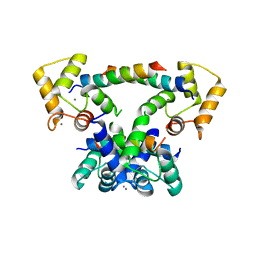 | | CaM-P458 complex (crystal form 2) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6Y38
 
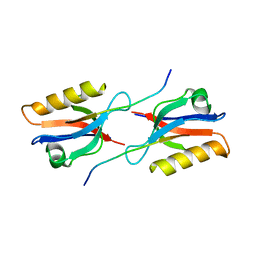 | | Crystal structure of Whirlin PDZ3 in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Chains: C,D, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-02-17 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9N
 
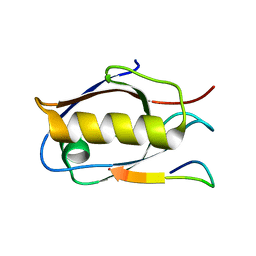 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Myosin 15a C-terminal PDZ binding motif peptide | | Descriptor: | Unconventional myosin-XV, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9O
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with CASK internal PDZ binding motif peptide | | Descriptor: | Peripheral plasma membrane protein CASK, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9Q
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Taperin internal PDZ binding motif peptide | | Descriptor: | Taperin, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.315 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
6Y9P
 
 | | Crystal structure of Whirlin PDZ3_C-ter in complex with Harmonin a1 C-terminal PDZ binding motif peptide | | Descriptor: | Harmonin a1, Whirlin | | Authors: | Zhu, Y, Delhommel, F, Haouz, A, Caillet-Saguy, C, Vaney, M, Mechaly, A.E, Wolff, N. | | Deposit date: | 2020-03-10 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.169 Å) | | Cite: | Deciphering the Unexpected Binding Capacity of the Third PDZ Domain of Whirlin to Various Cochlear Hair Cell Partners.
J.Mol.Biol., 432, 2020
|
|
7QBJ
 
 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QDX
 
 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QEM
 
 | | bacterial IMPDH chimera | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Gedeon, A, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2021-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
8P5T
 
 | | Single particle cryo-EM structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5V
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum in complex with the product succinyl-CoA | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5R
 
 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5U
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum with Coenzyme A bound to the E2o domain | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, COENZYME A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5X
 
 | | Single particle cryo-EM structure of the complex between Corynebacterium glutamicum homohexameric 2-oxoglutarate dehydrogenase OdhA and the FHA-protein inhibitor OdhI | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, MAGNESIUM ION, Oxoglutarate dehydrogenase inhibitor, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8P5W
 
 | | Single particle cryo-EM structure of homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum following reaction with the 2-oxoglutarate analogue succinyl phosphonate | | Descriptor: | (4~{S})-4-[(2~{R})-3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-2~{H}-1,3-thiazol-2-yl]-4-oxidanyl-4-phosphono-butanoic acid, 2-oxoglutarate dehydrogenase E1/E2 component, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Mechaly, A.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
6NQZ
 
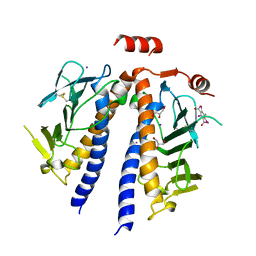 | | Flagellar protein FcpB from Leptospira interrogans | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Flagellar coiling protein B, GLYCEROL, ... | | Authors: | Trajtenberg, F, Larrieux, N, Buschiazzo, A. | | Deposit date: | 2019-01-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the Leptospira spirochete.
Elife, 9, 2020
|
|
7ZE0
 
 | | Solution structure of the PulM C-terminal domain from Klebsiella oxytoca | | Descriptor: | Type II secretion system protein M | | Authors: | Lopez-Castilla, A, Bardiaux, B, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2022-03-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
8P5S
 
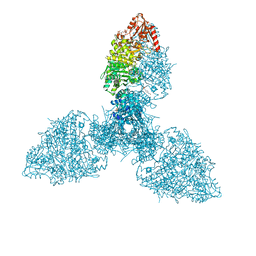 | | Crystal structure of the homohexameric 2-oxoglutarate dehydrogenase OdhA from Corynebacterium glutamicum | | Descriptor: | 2-oxoglutarate dehydrogenase E1/E2 component, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYL COENZYME *A, ... | | Authors: | Yang, L, Boyko, A, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
8PEK
 
 | |
7BES
 
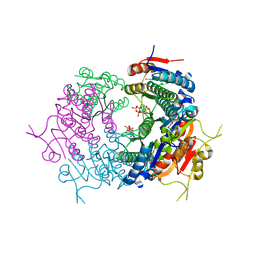 | | CryoEM structure of Mycobacterium tuberculosis UMP Kinase (UMPK) in complex with UDP and UTP | | Descriptor: | URIDINE 5'-TRIPHOSPHATE, URIDINE-5'-DIPHOSPHATE, Uridylate kinase | | Authors: | Bous, J, Trapani, S, Walter, P, Bron, P, Munier-Lehmann, H. | | Deposit date: | 2020-12-24 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the allosteric inhibition of UMP kinase from Gram-positive bacteria, a promising antibacterial target.
Febs J., 289, 2022
|
|
8P2V
 
 | | Neisseria meningitidis Type IV pilus SB-GATDH variant | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Neisseria meningitidis PilE variant SB-GATDH, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8P36
 
 | | Neisseria meningitidis Type IV pilus SB-DATDH variant | | Descriptor: | 2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, Neisseria meningitidis PilE, SB-DATDH variant, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8P3B
 
 | | Neisseria meningitidis Type IV pilus SA-GATDH variant | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Fimbrial protein, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8PIJ
 
 | | Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, C24 nanobody, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-21 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
