6LVB
 
 | | Structure of Dimethylformamidase, tetramer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LAY
 
 | |
6IFD
 
 | |
6IFI
 
 | |
6JOV
 
 | |
7W8J
 
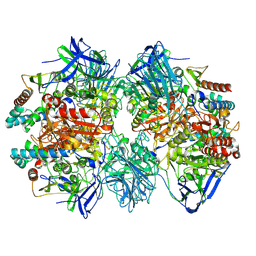 | | Dimethylformamidase, 2x(A2B2) | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Vinothkumar, K.R, Subramanian, R, Arya, C, Ramanathan, G. | | Deposit date: | 2021-12-07 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Dimethylformamidase with a Unique Iron Center
To Be Published
|
|
7VNL
 
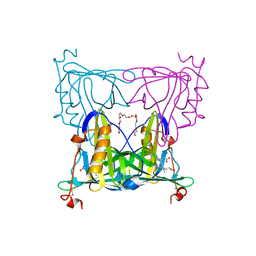 | | Sandercyanin mutant-F55A-Biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein, TETRAETHYLENE GLYCOL | | Authors: | Yadav, K, Ghosh, S, Subramanian, R. | | Deposit date: | 2021-10-11 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phenylalanine stacking enhances the red fluorescence of biliverdin IX alpha on UV excitation in sandercyanin fluorescent protein.
Febs Lett., 596, 2022
|
|
7VNS
 
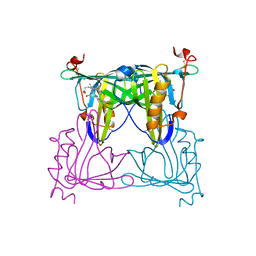 | | Sandercyanin mutant E79A-Biliverdin complex | | Descriptor: | BILIVERDINE IX ALPHA, Sandercyanin Fluorescent Protein | | Authors: | Yadav, K, Ghosh, S, Subramanian, R. | | Deposit date: | 2021-10-12 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phenylalanine stacking enhances the red fluorescence of biliverdin IX alpha on UV excitation in sandercyanin fluorescent protein.
Febs Lett., 596, 2022
|
|
5KQF
 
 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine (compound 12) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-4-methyl-6-pyrimidin-5-yl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1 | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
5KR8
 
 | | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine (compound 5) bound to BACE1 | | Descriptor: | (4~{S},6~{S})-4-[2,4-bis(fluoranyl)phenyl]-6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-methyl-5,6-dihydro-1,3-thiazin-2-amine, Beta-secretase 1, IODIDE ION | | Authors: | Lewis, H.A, Wu, Y.J, Rajamani, R, Thompson, L.A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Discovery of S3-Truncated, C-6 Heteroaryl Substituted Aminothiazine beta-Site APP Cleaving Enzyme-1 (BACE1) Inhibitors.
J.Med.Chem., 59, 2016
|
|
7BKX
 
 | | Diploptera punctata inspired lipocalin-like Milk protein expressed in Saccharomyces cerevisiae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Banerjee, S, Kanagavijayan, D, Subramanian, R, Santhakumari, P.R, Chavas, L.M.G, Ramaswamy, S. | | Deposit date: | 2021-01-17 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of recombinantly expressed cockroach Lili-Mip protein in glycosylated and deglycosylated forms.
Biochim Biophys Acta Gen Subj, 1866, 2022
|
|
6LVE
 
 | | Structure of Dimethylformamidase, tetramer, E521A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6LVC
 
 | | Structure of Dimethylformamidase, dimer | | Descriptor: | FE (III) ION, N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ZSX
 
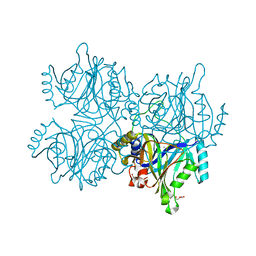 | | Catechol 2,3-dioxygenase with 3-fluorocatechol from Diaphorobacter sp DS2 | | Descriptor: | 1,2-ETHANEDIOL, 3-FLUOROBENZENE-1,2-DIOL, CALCIUM ION, ... | | Authors: | Mishra, K, Arya, C.K, Subramanian, R, Ramanathan, G. | | Deposit date: | 2018-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catechol 2,3-dioxygenase with 3-fluorocatechol from Diaphorobacter sp DS2.
To Be Published
|
|
5ZSZ
 
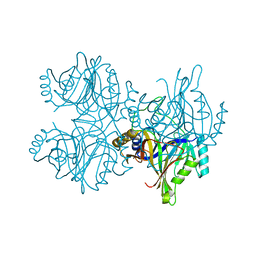 | | Catechol 2,3-dioxygenase (C23O64) from Diaphorobacter sp DS2 | | Descriptor: | CALCIUM ION, Catechol 2,3-dioxygenase, Extradiol ring cleavage protein, ... | | Authors: | Mishra, K, Arya, C.K, Subramanian, R, Ramanathan, G. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catechol 2,3-dioxygenase (C23O64) from Diaphorobacter sp DS2
To Be Published
|
|
6LVD
 
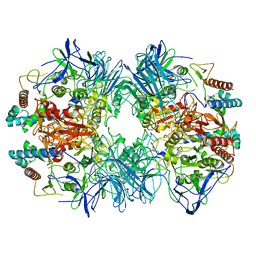 | | Structure of Dimethylformamidase, tetramer, Y440A mutant | | Descriptor: | N,N-dimethylformamidase large subunit, N,N-dimethylformamidase small subunit | | Authors: | Arya, C.A, Yadav, S, Fine, J, Casanal, A, Chopra, G, Ramanathan, G, Subramanian, R, Vinothkumar, K.R. | | Deposit date: | 2020-02-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A 2-Tyr-1-carboxylate Mononuclear Iron Center Forms the Active Site of a Paracoccus Dimethylformamidase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5ZA4
 
 | |
5ZNH
 
 | | Catechol 2,3-dioxygenase with 4-methyl catechol from Diaphorobacter sp DS2 | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLCATECHOL, CALCIUM ION, ... | | Authors: | Mishra, K, Arya, C.K, Subramaniyan, R, Ramanathan, G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | catechol 2,3-dioxygenase with 4-methyl catechol from Diaphorobacter sp DS2
To Be Published
|
|
1IHB
 
 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
6QTJ
 
 | | Crystal structure of human CDK8/CYCC in complex with BI 919811 | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, ~{N},~{N}-dimethyl-2-[4-[4-(2,6-naphthyridin-4-yl)phenyl]pyrazol-1-yl]ethanamide | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
6QTG
 
 | | Crystal structure of human CDK8/CYCC in complex with BI-1347 | | Descriptor: | 2-[4-(4-isoquinolin-4-ylphenyl)pyrazol-1-yl]-~{N},~{N}-dimethyl-ethanamide, Cyclin-C, Cyclin-dependent kinase 8 | | Authors: | Boettcher, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
6R3S
 
 | | CRYSTAL STRUCTURE OF CDK8-CycC IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[5-chloranyl-4-[(1~{S})-1-oxidanylethyl]pyridin-3-yl]-3,4-dihydro-2~{H}-1,8-naphthyridine-1-carboxamide, Cyclin-C, ... | | Authors: | Boettcher, J. | | Deposit date: | 2019-03-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Selective and Potent CDK8/19 Inhibitors Enhance NK-Cell Activity and Promote Tumor Surveillance.
Mol.Cancer Ther., 19, 2020
|
|
4GMX
 
 | |
2KAD
 
 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
7XNZ
 
 | | Native cystathionine beta-synthase of Mycobacterium tuberculosis. | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative cystathionine beta-synthase Rv1077 | | Authors: | Bandyopadhyay, P, Pramanick, I, Biswas, R, Sabarinath, P.S, Sreedharan, S, Singh, S, Rajmani, R, Laxman, S, Dutta, S, Singh, A. | | Deposit date: | 2022-04-30 | | Release date: | 2022-05-25 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | S-Adenosylmethionine-responsive cystathionine beta-synthase modulates sulfur metabolism and redox balance in Mycobacterium tuberculosis.
Sci Adv, 8, 2022
|
|
