1YIX
 
 | | Crystal structure of YCFH, TATD homolog from Escherichia coli K12, at 1.9 A resolution | | Descriptor: | ZINC ION, deoxyribonuclease ycfH | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-13 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ycfH, tatD homolog from Escherichia coli
To be Published
|
|
1YVQ
 
 | | The low salt (PEG) crystal structure of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5) | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of CO Hemoglobin E (betaE26K) approaching physiological pH (pH 7.5)
To be Published
|
|
1XWY
 
 | | Crystal structure of tatD deoxyribonuclease from Escherichia coli K12 at 2.0 A resolution | | Descriptor: | Deoxyribonuclease tatD, ZINC ION | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-11-02 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tatD DNase from Escherichia coli at 2.0 A resolution
To be Published
|
|
1ZZM
 
 | | Crystal structure of YJJV, TATD Homolog from Escherichia coli k12, at 1.8 A resolution | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ZINC ION, putative deoxyribonuclease yjjV | | Authors: | Malashkevich, V.N, Xiang, D.F, Raushel, F.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-06-14 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of YJJV, TATD homolog from Escherichia coli K12, at 1.8 A resolution
To be Published
|
|
1YVT
 
 | | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35) | | Descriptor: | CARBON MONOXIDE, GLYCEROL, Hemoglobin alpha chain, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2005-02-16 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The high salt (phosphate) crystal structure of CO Hemoglobin E (Glu26Lys) at physiological pH (pH 7.35)
To be Published
|
|
1AKB
 
 | |
1AKA
 
 | |
1AKC
 
 | |
2IJQ
 
 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
3QK7
 
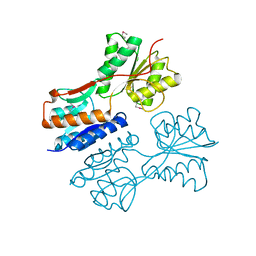 | | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001 | | Descriptor: | Transcriptional regulators | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of putative Transcriptional regulator from Yersinia pestis biovar Microtus str. 91001
To be Published
|
|
3RMT
 
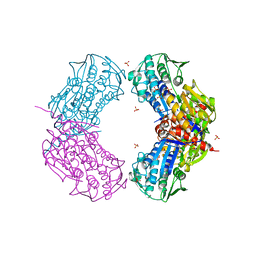 | | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125 | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase 1, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Ramagopal, U, Zencheck, W, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate synthase from Bacillus halodurans C-125
To be Published
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3RHD
 
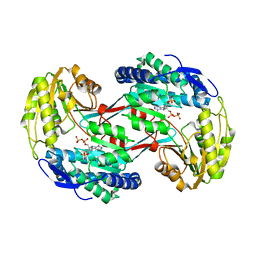 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP | | Descriptor: | Lactaldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 complexed with NADP
To be Published
|
|
3RHH
 
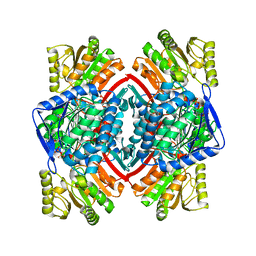 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
3UOG
 
 | | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Alcohol dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative Alcohol dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3UIF
 
 | | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein SsuA from Methylobacillus flagellatus KT | | Descriptor: | GLYCEROL, SULFATE ION, Sulfonate ABC transporter, ... | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-04 | | Release date: | 2011-11-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative sulfonate ABC transporter, periplasmic sulfonate-binding protein
SsuA from Methylobacillus flagellatus KT
To be Published
|
|
1MAP
 
 | |
1MAQ
 
 | |
3K4U
 
 | | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes DSM 1740 complexed with lysine | | Descriptor: | BINDING COMPONENT OF ABC TRANSPORTER, LYSINE | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-03 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | CRYSTAL STRUCTURE OF putative binding component of ABC transporter from Wolinella succinogenes
DSM 1740 complexed with lysine
To be Published
|
|
3K4I
 
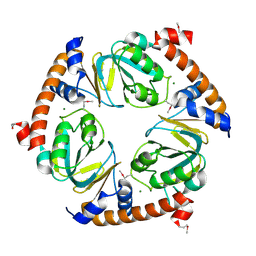 | | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, uncharacterized protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-10-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | CRYSTAL STRUCTURE OF uncharacterized protein PSPTO_3204 from Pseudomonas syringae pv. tomato
str. DC3000
To be Published
|
|
3K4H
 
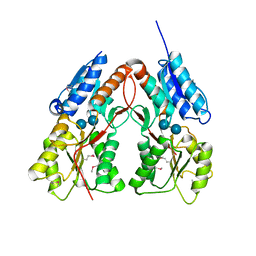 | | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, putative transcriptional regulator | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-10 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | CRYSTAL STRUCTURE OF putative transcriptional regulator LacI from Bacillus cereus subsp. cytotoxis NVH 391-98
To be Published
|
|
3K4W
 
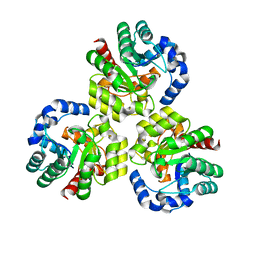 | | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein Bb4693 From Bordetella Bronchiseptica | | Descriptor: | uncharacterized protein Bb4693 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-06 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | CRYSTAL STRUCTURE OF Uncharacterized Tim-Barrel Protein
Bb4693 From Bordetella Bronchiseptica
To be Published
|
|
3L6D
 
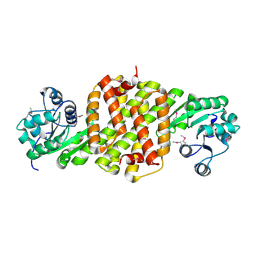 | | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440 | | Descriptor: | Putative oxidoreductase | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-12 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of putative oxidoreductase from Pseudomonas putida KT2440
To be Published
|
|
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3L6E
 
 | | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas hydrophila subsp. hydrophila ATCC 7966 | | Descriptor: | Oxidoreductase, short-chain dehydrogenase/reductase family, SULFATE ION | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-12-23 | | Release date: | 2010-02-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative short chain dehydrogenase/reductase family oxidoreductase from Aeromonas
hydrophila subsp. hydrophila ATCC 7966
To be Published
|
|
