8EGN
 
 | |
8EGM
 
 | |
8EGL
 
 | |
8EWA
 
 | |
3MD0
 
 | |
6HCW
 
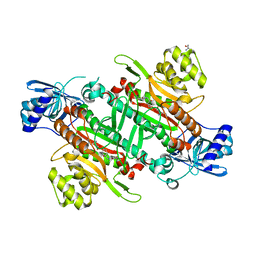 | | Crystal Structure of Lysyl-tRNA Synthetase from Cryptosporidium parvum complexed with L-lysine and a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HCV
 
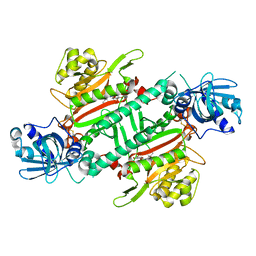 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with a chromone ligand | | Descriptor: | 6-fluoranyl-~{N}-[(1-oxidanylcyclohexyl)methyl]-4-oxidanylidene-chromene-2-carboxamide, Lysine--tRNA ligase | | Authors: | Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6HCU
 
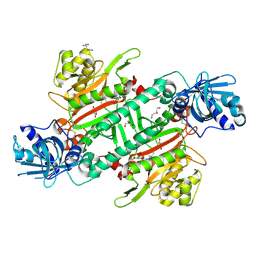 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum bound to a difluoro cyclohexyl chromone ligand | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HISTIDINE, ... | | Authors: | Tamjar, J, Robinson, D.A, Baragana, B, Norcross, N, Forte, B, Walpole, C, Gilbert, I.H. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4DZ3
 
 | |
3NRR
 
 | |
5BR9
 
 | |
5BT8
 
 | |
5BNZ
 
 | |
5BQ2
 
 | |
7SKB
 
 | |
7T35
 
 | |
7SZP
 
 | |
7SY9
 
 | |
7TCM
 
 | |
7T7J
 
 | |
7SBC
 
 | |
7SIR
 
 | |
7SBJ
 
 | |
7SIQ
 
 | |
5EJ2
 
 | |
