5M62
 
 | | Structure of the Mus musclus Langerin carbohydrate recognition domain in complex with glucose | | Descriptor: | C-type lectin domain family 4 member K, CALCIUM ION, GLYCEROL, ... | | Authors: | Loll, B, Aretz, J, Rademacher, C, Wahl, M.C. | | Deposit date: | 2016-10-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bacterial Polysaccharide Specificity of the Pattern Recognition Receptor Langerin Is Highly Species-dependent.
J. Biol. Chem., 292, 2017
|
|
7ALO
 
 | | Structure of B*27:09/photoRL9 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loll, B, Lan, H, Freund, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules.
Nat Commun, 12, 2021
|
|
7B3A
 
 | | Crystal structure of PamZ | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Loll, B, Dang, T, Mainz, A, Suessmuth, R, Wahl, M.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Molecular basis of antibiotic self-resistance in a bee larvae pathogen.
Nat Commun, 13, 2022
|
|
1W5C
 
 | | Photosystem II from Thermosynechococcus elongatus | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, CHLOROPHYLL A, ... | | Authors: | Biesiadka, J, Loll, B, Kern, J, Irrgang, K.-D, Saenger, W. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of Cyanobacterial Photosystem II at 3.2 A Resolution: A Closer Look at the Mn- Cluster
Phys.Chem.Chem.Phys., 6, 2004
|
|
6ET8
 
 | | Crystal structure of AlbA in complex with albicidin | | Descriptor: | Albicidin resistance protein, SULFATE ION, albicidin | | Authors: | Driller, R, Rostock, L, Alings, C, Graetz, S, Suessmuth, R, Mainz, A, Wahl, M.C, Loll, B. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular insights into antibiotic resistance - how a binding protein traps albicidin.
Nat Commun, 9, 2018
|
|
3D9J
 
 | | Snapshots of the RNA processing factor SCAF8 bound to different phosphorylated forms of the Carboxy-Terminal Domain of RNA-Polymerase II | | Descriptor: | AMMONIUM ION, GLYCEROL, RNA-binding protein 16, ... | | Authors: | Becker, R, Loll, B, Meinhart, A. | | Deposit date: | 2008-05-27 | | Release date: | 2008-06-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Snapshots of the RNA Processing Factor SCAF8 Bound to Different Phosphorylated Forms of the Carboxyl-terminal Domain of RNA Polymerase II.
J.Biol.Chem., 283, 2008
|
|
3D9N
 
 | |
7PLQ
 
 | | Crystal structure of the PARP domain of wheat SRO1 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Wirthmueller, L, Loll, B. | | Deposit date: | 2021-09-01 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The superior salinity tolerance of bread wheat cultivar Shanrong No. 3 is unlikely to be caused by elevated Ta-sro1 poly-(ADP-ribose) polymerase activity.
Plant Cell, 34, 2022
|
|
6TM8
 
 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 4 | | Descriptor: | Envelope glycoprotein D, GLYCEROL | | Authors: | Kremling, V, Loll, B, Osterrieder, N, Wahl, M, Dahmani, I, Chiantia, P, Azab, W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
6TEO
 
 | | Crystal structure of a yeast Snu114-Prp8 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | Authors: | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQO
 
 | | rrn anti-termination complex | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6Q9M
 
 | | Central Fibronectin-III array of RIM-binding protein | | Descriptor: | PHOSPHATE ION, RIM-binding protein, isoform F | | Authors: | Driller, J.D, Habibi, S, Wahl, M.C, Loll, B. | | Deposit date: | 2018-12-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | RIM-binding protein couples synaptic vesicle recruitment to release sites.
J.Cell Biol., 219, 2020
|
|
6ZCA
 
 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (monomer) | | Descriptor: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
7PH1
 
 | | Trypsin in complex with BPTI mutant (2S)-2-amino-4-monofluorobutanoic acid | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Dimos, N, Leppkes, J, Koksch, B, Loll, B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Water Network in the Binding Pocket of Fluorinated BPTI-Trypsin Complexes─Insights from Simulation and Experiment.
J.Phys.Chem.B, 126, 2022
|
|
2A83
 
 | | Crystal structure of hla-b*2705 complexed with the glucagon receptor (gr) peptide (residues 412-420) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Ruckert, C, Fiorillo, M.T, Loll, B, Moretti, R, Biesiadka, J, Saenger, W, Ziegler, A, Sorrentino, R, Uchanska-Ziegler, B. | | Deposit date: | 2005-07-07 | | Release date: | 2005-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype.
J.Biol.Chem., 281, 2006
|
|
6SQJ
 
 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein D, ... | | Authors: | Kremling, V, Loll, B, Azab, W, Osterrieder, N, Dahmani, I, Chiantia, P, Wahl, M. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
6Z9T
 
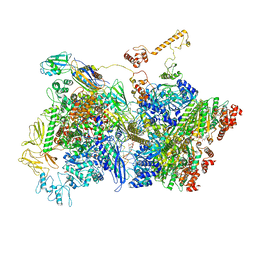 | | Transcription termination intermediate complex 5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9P
 
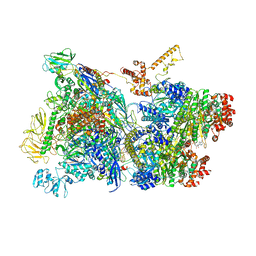 | | Transcription termination intermediate complex 1 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9Q
 
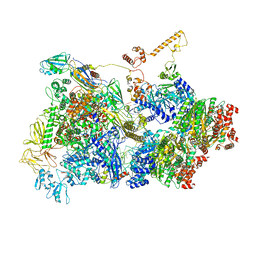 | | Transcription termination intermediate complex 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
7Z3S
 
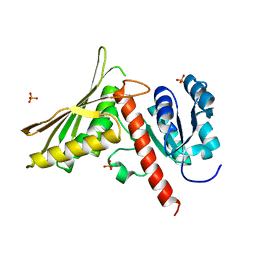 | |
6Z9S
 
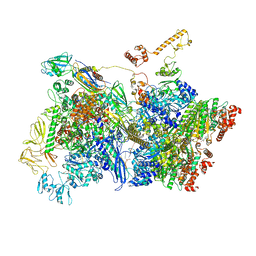 | | Transcription termination intermediate complex 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9R
 
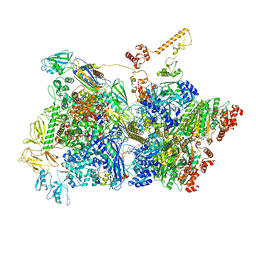 | | Transcription termination intermediate complex 3 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
9FMQ
 
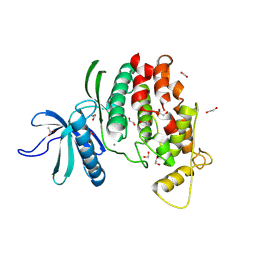 | | Crystal structure of C. merolae LAMMER-like dual specificity kinase (CmLIK) hairpin mutant kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dimos-Roehl, B, Haltenhof, T, Kotte, A, Heyd, F, Loll, B. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of C. merolae LAMMER-like dual specificity kinase CmLIK kinase domain
to be published
|
|
9FMP
 
 | | Crystal structure of phosphorylated C. merolae LAMMER-like dual specificity kinase (CmLIK) kinase domain in complex with adenosine | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, LAMMER-like dual specificity kinase | | Authors: | Dimos-Roehl, B, Haltenhof, T, Kotte, A, Heyd, F, Loll, B. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of C. merolae LAMMER-like dual specificity kinase CmLIK kinase domain
to be published
|
|
