7UB5
 
 | |
3TMP
 
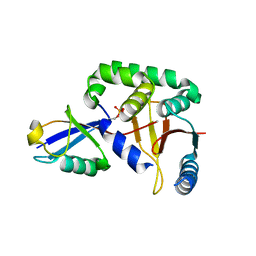 | | The catalytic domain of human deubiquitinase DUBA in complex with ubiquitin aldehyde | | Descriptor: | OTU domain-containing protein 5, Polyubiquitin-C | | Authors: | Ma, X, Yin, J, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3TMO
 
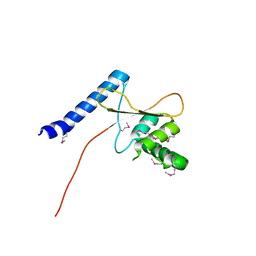 | | The catalytic domain of human deubiquitinase DUBA | | Descriptor: | OTU domain-containing protein 5 | | Authors: | Yin, J, Bosanac, I, Ma, X, Hymowitz, S, Starovasnik, M, Cochran, A. | | Deposit date: | 2011-08-31 | | Release date: | 2012-01-11 | | Last modified: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent activity of the deubiquitinase DUBA.
Nat.Struct.Mol.Biol., 19, 2012
|
|
