4AXP
 
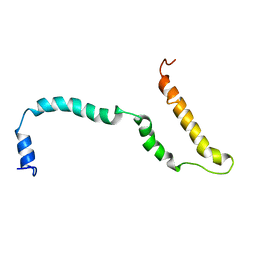 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
2IGG
 
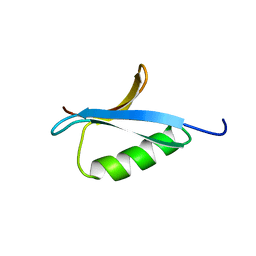 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
2IGH
 
 | | DETERMINATION OF THE SOLUTION STRUCTURES OF DOMAINS II AND III OF PROTEIN G FROM STREPTOCOCCUS BY 1H NMR | | Descriptor: | PROTEIN G | | Authors: | Lian, L.-Y, Derrick, J.P, Sutcliffe, M.J, Yang, J.C, Roberts, G.C.K. | | Deposit date: | 1992-08-26 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of domains II and III of protein G from Streptococcus by 1H nuclear magnetic resonance.
J.Mol.Biol., 228, 1992
|
|
1B1C
 
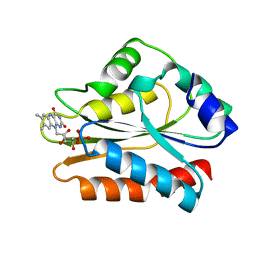 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
5AEQ
 
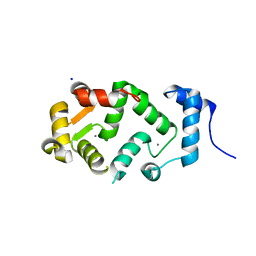 | | Neuronal calcium sensor (NCS-1)from Rattus norvegicus | | Descriptor: | CALCIUM ION, NEURONAL CALCIUM SENSOR 1, SODIUM ION | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5AER
 
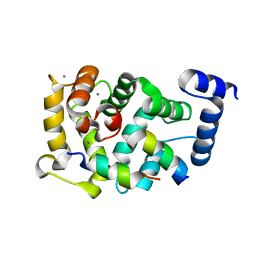 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
5MY7
 
 | | Adhesin Complex Protein from Neisseria meningitidis | | Descriptor: | Adhesin, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Derrick, J.P, Awanye, A. | | Deposit date: | 2017-01-25 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor.
PLoS Pathog., 13, 2017
|
|
1IMP
 
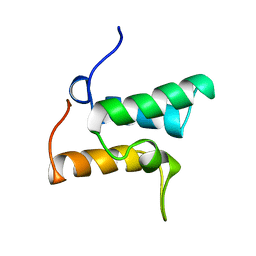 | | COLICIN E9 IMMUNITY PROTEIN IM9, NMR, 21 STRUCTURES | | Descriptor: | IM9 | | Authors: | Osborne, M.J, Breeze, A.L, Lian, L.-Y, Reilly, A, James, R, Kleanthous, C, Moore, G.R. | | Deposit date: | 1996-05-30 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and 13C nuclear magnetic resonance assignments of the colicin E9 immunity protein Im9.
Biochemistry, 35, 1996
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
2IVW
 
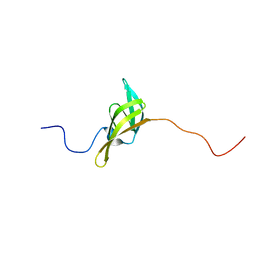 | | The solution structure of a domain from the Neisseria meningitidis PilP pilot protein. | | Descriptor: | PILP PILOT PROTEIN | | Authors: | Golovanov, A.P, Balasingham, S, Tzitzilonis, C, Goult, B.T, Lian, L.-Y, Homberset, H, Tonjum, T, Derrick, J.P. | | Deposit date: | 2006-06-20 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a domain from the Neisseria meningitidis lipoprotein PilP reveals a new beta-sandwich fold.
J. Mol. Biol., 364, 2006
|
|
1CC0
 
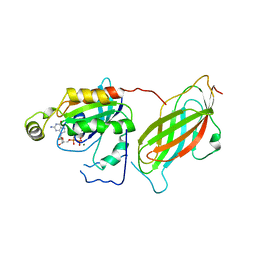 | | CRYSTAL STRUCTURE OF THE RHOA.GDP-RHOGDI COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, rho GDP dissociation inhibitor alpha, ... | | Authors: | Longenecker, K.L, Read, P, Derewenda, U, Dauter, Z, Garrard, S, Walker, L, Somlyo, A.V, Somlyo, A.P, Nakamoto, R.K, Derewenda, Z.S. | | Deposit date: | 1999-03-03 | | Release date: | 2000-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | How RhoGDI binds Rho.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6YWZ
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[(~{E})-(4-oxidanylidenebutanoylhydrazinylidene)methyl]benzoic acid, ARGININE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6YX1
 
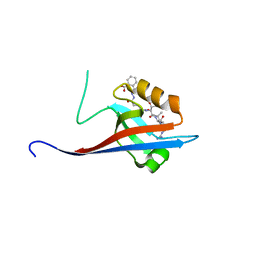 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[[2-(5-oxidanylidenepentanoyl)hydrazinyl]methyl]benzoic acid, ARGININE, LEUCINE, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6YX2
 
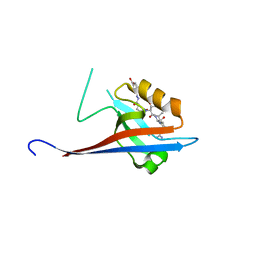 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[(~{E})-5-oxidanylidenepentanoyldiazenyl]methyl]benzoic acid, PWW-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
2I38
 
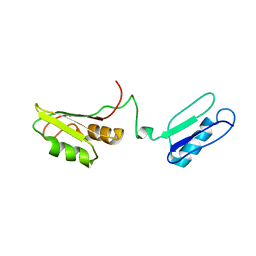 | | Solution structure of the RRM of SRp20 | | Descriptor: | Fusion protein consists of immunoglobulin G-Binding Protein G and Splicing factor, arginine/serine-rich 3 | | Authors: | Hargous, Y.F, Allain, F.H.T. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
2I2Y
 
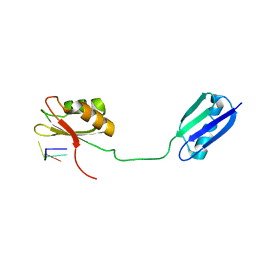 | | Solution structure of the RRM of SRp20 bound to the RNA CAUC | | Descriptor: | (5'-R(*CP*AP*UP*C)-3'), Fusion protein consists of immunoglobulin G-Binding Protein G and Splicing factor, arginine/serine-rich 3 | | Authors: | Hargous, Y.F, Allain, F.H. | | Deposit date: | 2006-08-17 | | Release date: | 2006-12-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8.
Embo J., 25, 2006
|
|
6XY3
 
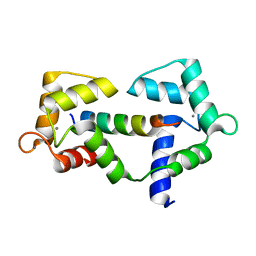 | |
6XXF
 
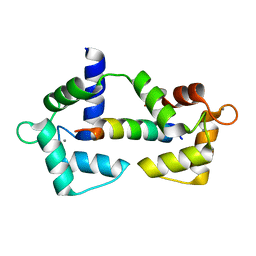 | |
6XXX
 
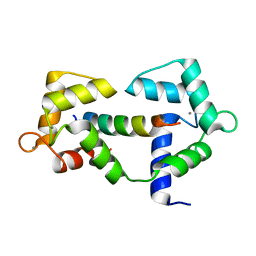 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
6YX0
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[2-(4-oxidanylidenebutanoyl)hydrazinyl]methyl]benzoic acid, PWQ-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
1IGD
 
 | |
2LNT
 
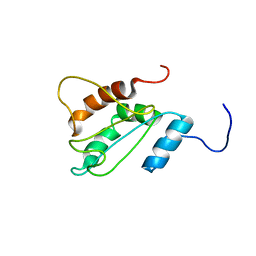 | | Solution structure of E60A mutant AGR2 | | Descriptor: | Anterior gradient protein 2 homolog | | Authors: | Patel, P, Clarke, C.J, Barraclough, D.L, Rudland, P.S, Barraclough, R, Lian, L. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metastasis-promoting anterior gradient 2 protein has a dimeric thioredoxin fold structure and a role in cell adhesion.
J.Mol.Biol., 425, 2013
|
|
2LNS
 
 | | Solution structure of AGR2 residues 41-175 | | Descriptor: | Anterior gradient protein 2 homolog | | Authors: | Patel, P, Clarke, C.J, Barraclough, D.L, Rudland, P.S, Barraclough, R, Lian, L. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Metastasis-promoting anterior gradient 2 protein has a dimeric thioredoxin fold structure and a role in cell adhesion.
J.Mol.Biol., 425, 2013
|
|
2LV7
 
 | | Solution structure of Ca2+-bound CaBP7 N-terminal doman | | Descriptor: | CALCIUM ION, Calcium-binding protein 7 | | Authors: | Mccue, H.V, Patel, P, Herbert, A.P, Lian, L, Burgoyne, R.D, Haynes, L.P. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ca2+-bound N-terminal Domain of CaBP7: A REGULATOR OF GOLGI TRAFFICKING.
J.Biol.Chem., 287, 2012
|
|
