8D1C
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-(Trifluoromethoxy)benzoic acid | | Descriptor: | 4-(trifluoromethoxy)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-05-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Oxidations Using a Cytochrome P450 Enzyme Variant Driven with Surrogate Oxygen Donors and Light.
Chemistry, 28, 2022
|
|
8GLZ
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid. Crystal was initially co-crystallised with 4-methoxybenzoic acid and soaked with 4 mM hydrogen peroxide | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM2
 
 | |
8GLY
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM1
 
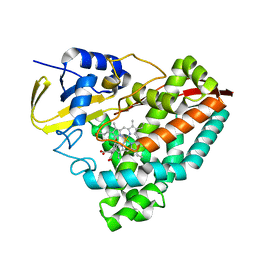 | | Crystal structure of T252E-CYP199A4 in complex with 4-methoxybenzoic acid soaked with 1 mM hydrogen peroxide | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
1HOX
 
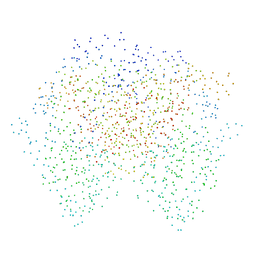 | | CRYSTAL STRUCTURE OF RABBIT PHOSPHOGLUCOSE ISOMERASE COMPLEXED WITH FRUCTOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Jeffrey, C.J, Lee, J.H, Chang, K.Z, Patel, V. | | Deposit date: | 2000-12-11 | | Release date: | 2001-07-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate D-fructose 6-phosphate.
Biochemistry, 40, 2001
|
|
5WQ0
 
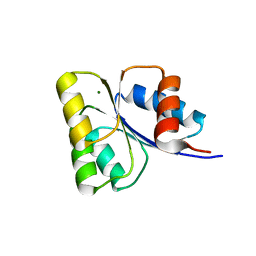 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
5YL7
 
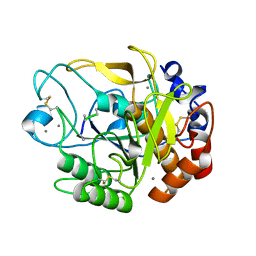 | | Proteases from Pseudoalteromonas arctica PAMC 21717 (Pro21717) | | Descriptor: | CALCIUM ION, Copurified unknown peptide, Pseudoalteromonas arctica PAMC 21717 | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2017-10-17 | | Release date: | 2018-01-31 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 angstrom resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme
PLoS ONE, 13, 2018
|
|
5XGW
 
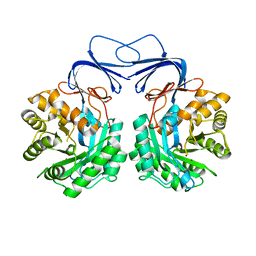 | |
5Z2E
 
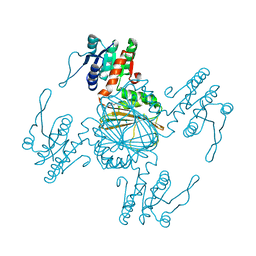 | |
5Z2F
 
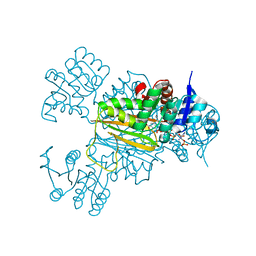 | | NADPH/PDA bound Dihydrodipicolinate reductase from Paenisporosarcina sp. TG-14 | | Descriptor: | Dihydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase (PaDHDPR) from Paenisporosarcina sp. TG-14: structural basis for NADPH preference as a cofactor
Sci Rep, 8, 2018
|
|
7NFX
 
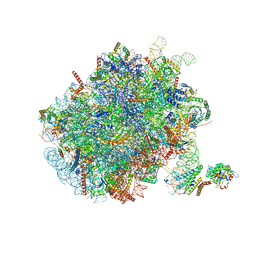 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
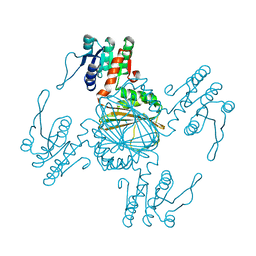
5Z2D
 
 | |
5XGX
 
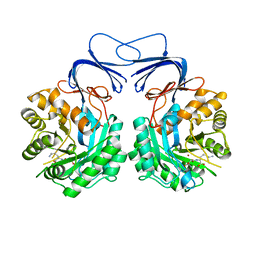 | | Crystal structure of colwellia psychrerythraea strain 34H isoaspartyl dipeptidase E80Q mutant complexed with beta-isoaspartyl lysine | | Descriptor: | D-ASPARTIC ACID, D-LYSINE, Isoaspartyl dipeptidase, ... | | Authors: | Lee, J.H, Lee, C.W, Park, S.H. | | Deposit date: | 2017-04-18 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure and functional characterization of an isoaspartyl dipeptidase (CpsIadA) from Colwellia psychrerythraea strain 34H.
PLoS ONE, 12, 2017
|
|
5BVS
 
 | | Linoleate-bound pFABP4 | | Descriptor: | Fatty acid-binding protein, LINOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVQ
 
 | | Ligand-unbound pFABP4 | | Descriptor: | fatty acid-binding protein | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
5BVT
 
 | | Palmitate-bound pFABP5 | | Descriptor: | Epidermal fatty acid-binding protein, PALMITOLEIC ACID | | Authors: | Lee, J.H, Lee, C.W, Do, H. | | Deposit date: | 2015-06-05 | | Release date: | 2015-08-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the ligand-binding specificity of fatty acid-binding proteins (pFABP4 and pFABP5) in gentoo penguin
Biochem.Biophys.Res.Commun., 465, 2015
|
|
4NR0
 
 | |
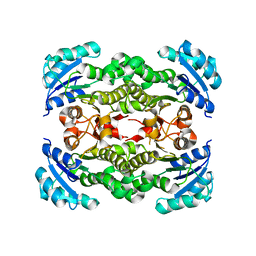
4NQZ
 
 | |
5UY3
 
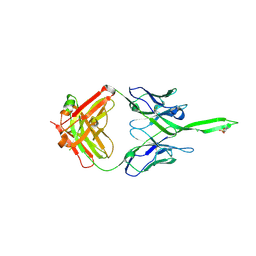 | | Crystal structure of human Fab PGT144, a broadly reactive and potent HIV-1 neutralizing antibody | | Descriptor: | Antibody PGT144 Fab heavy chain, Antibody PGT144 Fab light chain | | Authors: | Julien, J.-P, Lee, J.H, Wilson, I.A. | | Deposit date: | 2017-02-23 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
5BY2
 
 | |
7C4X
 
 | |
6JQS
 
 | | Structure of Transcription factor, GerE | | Descriptor: | DNA-binding response regulator | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2019-04-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a transcription factor, GerE (PaGerE), from spore-forming bacterium Paenisporosarcina sp. TG-14.
Biochem.Biophys.Res.Commun., 513, 2019
|
|
6IFH
 
 | | Unphosphorylated Spo0F from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase F | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unphosphorylated Spo0F from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Biodesign, 6(4), 2019
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
