3BNN
 
 | |
3BNS
 
 | |
3BNR
 
 | |
3BNO
 
 | |
3BNL
 
 | |
3BNQ
 
 | |
5XUV
 
 | | Crystal structure of DNA duplex containing 4-thiothymine-2Ag(I)-4-thiothymine base pairs | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(8RO)P*(8RO)P*TP*CP*GP*CP*G)-3'), POTASSIUM ION, SILVER ION | | Authors: | Kondo, J, Sugawara, T, Saneyoshi, H, Ono, A. | | Deposit date: | 2017-06-26 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a DNA duplex containing four Ag(i) ions in consecutive dinuclear Ag(i)-mediated base pairs: 4-thiothymine-2Ag(i)-4-thiothymine
Chem. Commun. (Camb.), 53, 2017
|
|
1UHY
 
 | | Crystal structure of d(GCGATAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*TP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
1UHX
 
 | | Crystal structure of d(GCGAGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Umeda, S.I, Fujita, K, Sunami, T, Takenaka, A. | | Deposit date: | 2003-07-13 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray analyses of d(GCGAXAGC) containing G and T at X: the base-intercalated duplex is still stable even in point mutants at the fifth residue.
J.Synchrotron Radiat., 11, 2004
|
|
7YVX
 
 | |
1V3O
 
 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
4P3U
 
 | |
4P3S
 
 | |
4P3T
 
 | |
1V3N
 
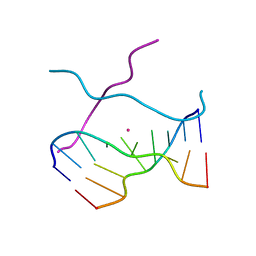 | | Crystal structure of d(GCGAGAGC): the DNA quadruplex structure split from the octaplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
1V3P
 
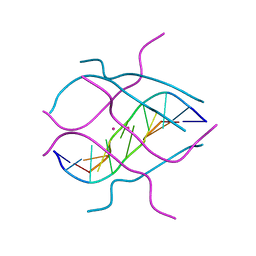 | | Crystal structure of d(GCGAGAGC): the DNA octaplex structure with I-motif of G-quartet | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*GP*AP*GP*C)-3', POTASSIUM ION | | Authors: | Kondo, J, Umeda, S, Sunami, T, Takenaka, A. | | Deposit date: | 2003-11-03 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a DNA octaplex with I-motif of G-quartets and its splitting into two quadruplexes suggest a folding mechanism of eight tandem repeats
Nucleic Acids Res., 32, 2004
|
|
4P43
 
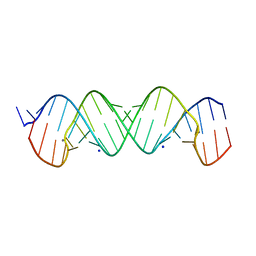 | |
4P20
 
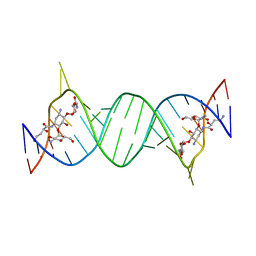 | | Crystal structures of the bacterial ribosomal decoding site complexed with amikacin | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Kondo, J, Francois, B, Russell, R.J.M, Murray, J.B, Westhof, E. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the gamma-amino-alpha-hydroxybutyryl (haba) group.
Biochimie, 88, 2006
|
|
3TD1
 
 | |
3TD0
 
 | |
2FQN
 
 | |
2FZA
 
 | | Crystal structure of d(GCGGGAGC): the base-intercalated duplex | | Descriptor: | 5'-D(*GP*(CBR)P*GP*GP*GP*AP*GP*C)-3', CALCIUM ION, SODIUM ION | | Authors: | Kondo, J, Ciengshin, T, Juan, E.C.M, Mitomi, K, Takenaka, A. | | Deposit date: | 2006-02-09 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of d(gcGXGAgc) with X=G: a mutation at X is possible to occur in a base-intercalated duplex for multiplex formation
Nucleosides Nucleotides Nucleic Acids, 25, 2006
|
|
2G5K
 
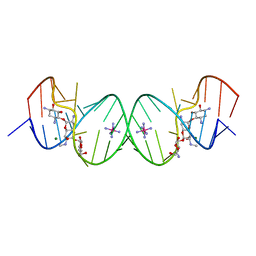 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with Apramycin | | Descriptor: | 5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3', APRAMYCIN, COBALT HEXAMMINE(III), ... | | Authors: | Kondo, J, Francois, B, Urzhumtsev, A, Westhof, E. | | Deposit date: | 2006-02-23 | | Release date: | 2006-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site Complexed with Apramycin
Angew.Chem.Int.Ed.Engl., 45, 2006
|
|
7EFG
 
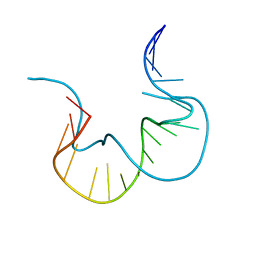 | | RNA kink-turn motif | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP*GP*AP*GP*CP*C)-3') | | Authors: | Kondo, J, Nagashima, M. | | Deposit date: | 2021-03-21 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | RNA kink-turn motif
To Be Published
|
|
2GOT
 
 | |
