3LR2
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Johansson, J, Knight, S.D, Rising, A, Casals, C, Saenz, A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LR6
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Saenz, A, Casals, C, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LR8
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Major ampullate spidroin 1, ... | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3LRD
 
 | | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay | | Descriptor: | 1,2-ETHANEDIOL, Major ampullate spidroin 1, TRIETHYLENE GLYCOL | | Authors: | Askarieh, G, Hedhammar, H, Nordling, K, Rising, A, Johansson, J, Knight, S.D, Saenz, A, Casals, C. | | Deposit date: | 2010-02-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Self-assembly of spider silk proteins is controlled by a pH-sensitive relay.
Nature, 465, 2010
|
|
3DOS
 
 | | Crystal structure of the complex of the Caf1M chaperone with the mini-fiber of two Caf1 subunits (Caf1:Caf1), carrying the Thr7Phe and Ala9Val mutations in the Gd donor strand | | Descriptor: | Chaperone protein caf1M, F1 capsule antigen | | Authors: | Fooks, L.J, Yu, X, Moslehi-Mohebi, E, Tischenko, V, Knight, S.D, MacIntyre, S, Zavialov, A.V. | | Deposit date: | 2008-07-06 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hydrophobicity and rigidity of binding segments enable CAF1M chaperone to act as assembly catalyst
TO BE PUBLISHED
|
|
3DPB
 
 | | Crystal structure of the complex of the Caf1M chaperone with the mini-fiber of two Caf1 subunits (Caf1:Caf1), carrying the Ala9Val, Ala11Val, and Leu13Val mutations in the Gd donor strand | | Descriptor: | Chaperone protein caf1M, F1 capsule antigen | | Authors: | Fooks, L.J, Yu, X, Moslehi-Mohebi, E, Tischenko, V, Knight, S.D, MacIntyre, S, Zavialov, A.V. | | Deposit date: | 2008-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrophobicity and rigidity of binding segments enable CAF1M chaperone to act as assembly catalyst
To be published
|
|
3DSN
 
 | | Crystal structure of the complex of the Caf1M chaperone with the mini-fiber of two Caf1 subunits (Caf1:Caf1), carrying the Thr7Phe mutation in the Gd donor strand | | Descriptor: | Chaperone protein caf1M, F1 capsule antigen | | Authors: | Fooks, L.J, Yu, X, Moslehi-Mohebi, E, Tischenko, V, Knight, S.D, MacIntyre, S, Zavialov, A.V. | | Deposit date: | 2008-07-13 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrophobicity and rigidity of binding segments enable CAF1M chaperone to act as assembly catalyst
To be Published
|
|
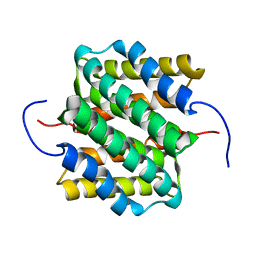
4FBS
 
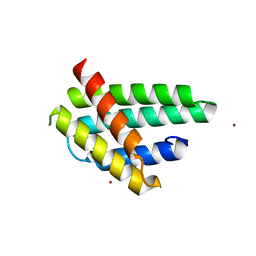 | | Structure of monomeric NT from Euprosthenops australis Major Ampullate Spidroin 1 (MaSp1) | | Descriptor: | BROMIDE ION, Major ampullate spidroin 1 | | Authors: | Askarieh, G, Hedhammar, M, Rising, A, Johansson, J, Knight, S.D. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain
J.Mol.Biol., 2012
|
|
2LPI
 
 | | NMR structure of a monomeric mutant (A72R) of major ampullate spidroin 1 N-terminal domain | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S.D, Johansson, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain.
J.Mol.Biol., 422, 2012
|
|
2LPJ
 
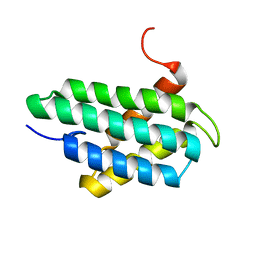 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 7.2 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S.D, Johansson, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Dimerization of Spider Silk N-Terminal Domain Requires Relocation of a Wedged Tryptophan Side Chain.
J.Mol.Biol., 422, 2012
|
|
3L54
 
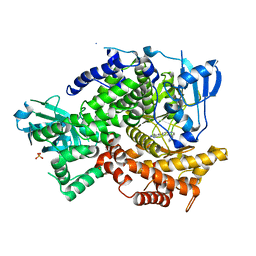 | | Structure of Pi3K gamma with inhibitor | | Descriptor: | 6-(1H-pyrazolo[3,4-b]pyridin-5-yl)-4-pyridin-4-ylquinoline, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Smallwood, A.M. | | Deposit date: | 2009-12-21 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
3L08
 
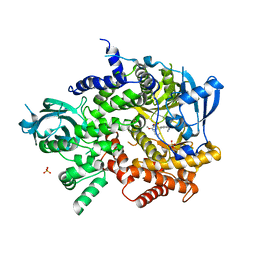 | | Structure of Pi3K gamma with a potent inhibitor: GSK2126458 | | Descriptor: | 2,4-difluoro-N-[2-methoxy-5-(4-pyridazin-4-ylquinolin-6-yl)pyridin-3-yl]benzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Elkins, P.A, Marrero, E.M. | | Deposit date: | 2009-12-09 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin.
ACS Med Chem Lett, 1, 2010
|
|
1QUN
 
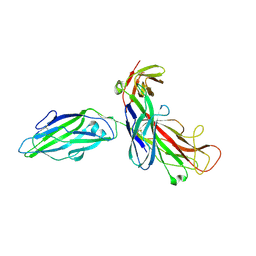 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
3FCG
 
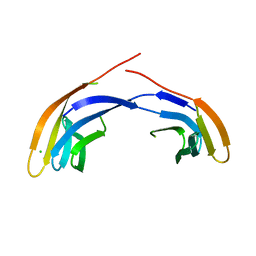 | | Crystal Structure Analysis of the Middle Domain of the Caf1A Usher | | Descriptor: | CHLORIDE ION, F1 capsule-anchoring protein | | Authors: | Yu, X, Visweswaran, G.R, Duck, Z, Marupakula, S, MacIntyre, S, Knight, S, Zavialov, A.V. | | Deposit date: | 2008-11-21 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Caf1A usher possesses a Caf1 subunit-like domain that is crucial for Caf1 fibre secretion
Biochem.J., 418, 2009
|
|
7ZL4
 
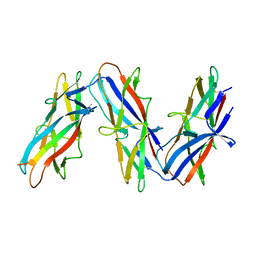 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | Descriptor: | CsuA/B | | Authors: | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
2LTH
 
 | | NMR structure of major ampullate spidroin 1 N-terminal domain at pH 5.5 | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Otikovs, M, Jaudzems, K, Nordling, K, Landreh, M, Rising, A, Askarieh, G, Knight, S, Johansson, J. | | Deposit date: | 2012-05-25 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation.
Nat Commun, 5, 2014
|
|
4YKN
 
 | | Pi3K alpha lipid kinase with Active Site Inhibitor | | Descriptor: | 3-(6-methoxypyridin-3-yl)-5-[({4-[(5-methyl-1,3,4-thiadiazol-2-yl)sulfamoyl]phenyl}amino)methyl]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha,Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform fusion protein | | Authors: | Elkins, P.A. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Potent Class of PI3K alpha Inhibitors with Unique Binding Mode via Encoded Library Technology (ELT).
Acs Med.Chem.Lett., 6, 2015
|
|
6FJY
 
 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8RNU
 
 | |
